Shenda Hong
National Institute of Health Data Science, Peking University, Beijing, China, Department of Emergency Medicine, Peking University First Hospital, Beijing, China
Aortic Valve Disease Detection from PPG via Physiology-Informed Self-Supervised Learning
Feb 04, 2026Abstract:Traditional diagnosis of aortic valve disease relies on echocardiography, but its cost and required expertise limit its use in large-scale early screening. Photoplethysmography (PPG) has emerged as a promising screening modality due to its widespread availability in wearable devices and its ability to reflect underlying hemodynamic dynamics. However, the extreme scarcity of gold-standard labeled PPG data severely constrains the effectiveness of data-driven approaches. To address this challenge, we propose and validate a new paradigm, Physiology-Guided Self-Supervised Learning (PG-SSL), aimed at unlocking the value of large-scale unlabeled PPG data for efficient screening of Aortic Stenosis (AS) and Aortic Regurgitation (AR). Using over 170,000 unlabeled PPG samples from the UK Biobank, we formalize clinical knowledge into a set of PPG morphological phenotypes and construct a pulse pattern recognition proxy task for self-supervised pre-training. A dual-branch, gated-fusion architecture is then employed for efficient fine-tuning on a small labeled subset. The proposed PG-SSL framework achieves AUCs of 0.765 and 0.776 for AS and AR screening, respectively, significantly outperforming supervised baselines trained on limited labeled data. Multivariable analysis further validates the model output as an independent digital biomarker with sustained prognostic value after adjustment for standard clinical risk factors. This study demonstrates that PG-SSL provides an effective, domain knowledge-driven solution to label scarcity in medical artificial intelligence and shows strong potential for enabling low-cost, large-scale early screening of aortic valve disease.
ECG-R1: Protocol-Guided and Modality-Agnostic MLLM for Reliable ECG Interpretation
Feb 04, 2026Abstract:Electrocardiography (ECG) serves as an indispensable diagnostic tool in clinical practice, yet existing multimodal large language models (MLLMs) remain unreliable for ECG interpretation, often producing plausible but clinically incorrect analyses. To address this, we propose ECG-R1, the first reasoning MLLM designed for reliable ECG interpretation via three innovations. First, we construct the interpretation corpus using \textit{Protocol-Guided Instruction Data Generation}, grounding interpretation in measurable ECG features and monograph-defined quantitative thresholds and diagnostic logic. Second, we present a modality-decoupled architecture with \textit{Interleaved Modality Dropout} to improve robustness and cross-modal consistency when either the ECG signal or ECG image is missing. Third, we present \textit{Reinforcement Learning with ECG Diagnostic Evidence Rewards} to strengthen evidence-grounded ECG interpretation. Additionally, we systematically evaluate the ECG interpretation capabilities of proprietary, open-source, and medical MLLMs, and provide the first quantitative evidence that severe hallucinations are widespread, suggesting that the public should not directly trust these outputs without independent verification. Code and data are publicly available at \href{https://github.com/PKUDigitalHealth/ECG-R1}{here}, and an online platform can be accessed at \href{http://ai.heartvoice.com.cn/ECG-R1/}{here}.
ECGFlowCMR: Pretraining with ECG-Generated Cine CMR Improves Cardiac Disease Classification and Phenotype Prediction
Jan 28, 2026Abstract:Cardiac Magnetic Resonance (CMR) imaging provides a comprehensive assessment of cardiac structure and function but remains constrained by high acquisition costs and reliance on expert annotations, limiting the availability of large-scale labeled datasets. In contrast, electrocardiograms (ECGs) are inexpensive, widely accessible, and offer a promising modality for conditioning the generative synthesis of cine CMR. To this end, we propose ECGFlowCMR, a novel ECG-to-CMR generative framework that integrates a Phase-Aware Masked Autoencoder (PA-MAE) and an Anatomy-Motion Disentangled Flow (AMDF) to address two fundamental challenges: (1) the cross-modal temporal mismatch between multi-beat ECG recordings and single-cycle CMR sequences, and (2) the anatomical observability gap due to the limited structural information inherent in ECGs. Extensive experiments on the UK Biobank and a proprietary clinical dataset demonstrate that ECGFlowCMR can generate realistic cine CMR sequences from ECG inputs, enabling scalable pretraining and improving performance on downstream cardiac disease classification and phenotype prediction tasks.
ECGomics: An Open Platform for AI-ECG Digital Biomarker Discovery
Jan 19, 2026Abstract:Background: Conventional electrocardiogram (ECG) analysis faces a persistent dichotomy: expert-driven features ensure interpretability but lack sensitivity to latent patterns, while deep learning offers high accuracy but functions as a black box with high data dependency. We introduce ECGomics, a systematic paradigm and open-source platform for the multidimensional deconstruction of cardiac signals into digital biomarker. Methods: Inspired by the taxonomic rigor of genomics, ECGomics deconstructs cardiac activity across four dimensions: Structural, Intensity, Functional, and Comparative. This taxonomy synergizes expert-defined morphological rules with data-driven latent representations, effectively bridging the gap between handcrafted features and deep learning embeddings. Results: We operationalized this framework into a scalable ecosystem consisting of a web-based research platform and a mobile-integrated solution (https://github.com/PKUDigitalHealth/ECGomics). The web platform facilitates high-throughput analysis via precision parameter configuration, high-fidelity data ingestion, and 12-lead visualization, allowing for the systematic extraction of biomarkers across the four ECGomics dimensions. Complementarily, the mobile interface, integrated with portable sensors and a cloud-based engine, enables real-time signal acquisition and near-instantaneous delivery of structured diagnostic reports. This dual-interface architecture successfully transitions ECGomics from theoretical discovery to decentralized, real-world health management, ensuring professional-grade monitoring in diverse clinical and home-based settings. Conclusion: ECGomics harmonizes diagnostic precision, interpretability, and data efficiency. By providing a deployable software ecosystem, this paradigm establishes a robust foundation for digital biomarker discovery and personalized cardiovascular medicine.
AnyECG: Evolved ECG Foundation Model for Holistic Health Profiling
Jan 12, 2026Abstract:Background: Artificial intelligence enabled electrocardiography (AI-ECG) has demonstrated the ability to detect diverse pathologies, but most existing models focus on single disease identification, neglecting comorbidities and future risk prediction. Although ECGFounder expanded cardiac disease coverage, a holistic health profiling model remains needed. Methods: We constructed a large multicenter dataset comprising 13.3 million ECGs from 2.98 million patients. Using transfer learning, ECGFounder was fine-tuned to develop AnyECG, a foundation model for holistic health profiling. Performance was evaluated using external validation cohorts and a 10-year longitudinal cohort for current diagnosis, future risk prediction, and comorbidity identification. Results: AnyECG demonstrated systemic predictive capability across 1172 conditions, achieving an AUROC greater than 0.7 for 306 diseases. The model revealed novel disease associations, robust comorbidity patterns, and future disease risks. Representative examples included high diagnostic performance for hyperparathyroidism (AUROC 0.941), type 2 diabetes (0.803), Crohn disease (0.817), lymphoid leukemia (0.856), and chronic obstructive pulmonary disease (0.773). Conclusion: The AnyECG foundation model provides substantial evidence that AI-ECG can serve as a systemic tool for concurrent disease detection and long-term risk prediction.
Case Prompting to Mitigate Large Language Model Bias for ICU Mortality Prediction
Dec 24, 2025Abstract:Accurate mortality risk prediction for intensive care unit (ICU) patients is essential for clinical decision-making. Although large language models (LLMs) show promise in predicting outcomes from structured medical data, their predictions may exhibit demographic biases related to sex, age, and race, limiting their trustworthy use in clinical practice. Existing debiasing methods often reduce predictive performance, making it difficult to jointly optimize fairness and accuracy. In this study, we systematically examine bias in LLM-based ICU mortality prediction and propose a training-free, clinically adaptive prompting framework to simultaneously improve fairness and performance. We first develop a multi-dimensional bias assessment scheme for comprehensive model diagnosis. Building on this analysis, we introduce CAse Prompting (CAP), a novel prompting framework that integrates conventional debiasing prompts with case-based reasoning. CAP guides the model to learn from similar historical misprediction cases and their correct outcomes, enabling correction of biased reasoning patterns. Experiments on the MIMIC-IV dataset show that CAP substantially improves both predictive accuracy and fairness. CAP increases AUROC from 0.806 to 0.873 and AUPRC from 0.497 to 0.694, while reducing sex- and race-related disparities by over 90%. Feature reliance analysis further indicates highly consistent attention patterns across demographic groups, with similarity scores exceeding 0.98. These results demonstrate that LLMs exhibit measurable bias in ICU mortality prediction, and that a carefully designed prompting framework can effectively co-optimize fairness and performance without retraining, offering a transferable paradigm for equitable clinical decision support.
Artificial Intelligence-Enabled Spirometry for Early Detection of Right Heart Failure
Nov 17, 2025Abstract:Right heart failure (RHF) is a disease characterized by abnormalities in the structure or function of the right ventricle (RV), which is associated with high morbidity and mortality. Lung disease often causes increased right ventricular load, leading to RHF. Therefore, it is very important to screen out patients with cor pulmonale who develop RHF from people with underlying lung diseases. In this work, we propose a self-supervised representation learning method to early detecting RHF from patients with cor pulmonale, which uses spirogram time series to predict patients with RHF at an early stage. The proposed model is divided into two stages. The first stage is the self-supervised representation learning-based spirogram embedding (SLSE) network training process, where the encoder of the Variational autoencoder (VAE-encoder) learns a robust low-dimensional representation of the spirogram time series from the data-augmented unlabeled data. Second, this low-dimensional representation is fused with demographic information and fed into a CatBoost classifier for the downstream RHF prediction task. Trained and tested on a carefully selected subset of 26,617 individuals from the UK Biobank, our model achieved an AUROC of 0.7501 in detecting RHF, demonstrating strong population-level distinction ability. We further evaluated the model on high-risk clinical subgroups, achieving AUROC values of 0.8194 on a test set of 74 patients with chronic kidney disease (CKD) and 0.8413 on a set of 64 patients with valvular heart disease (VHD). These results highlight the model's potential utility in predicting RHF among clinically elevated-risk populations. In conclusion, this study presents a self-supervised representation learning approach combining spirogram time series and demographic data, demonstrating promising potential for early RHF detection in clinical practice.
AnyECG-Lab: An Exploration Study of Fine-tuning an ECG Foundation Model to Estimate Laboratory Values from Single-Lead ECG Signals
Oct 25, 2025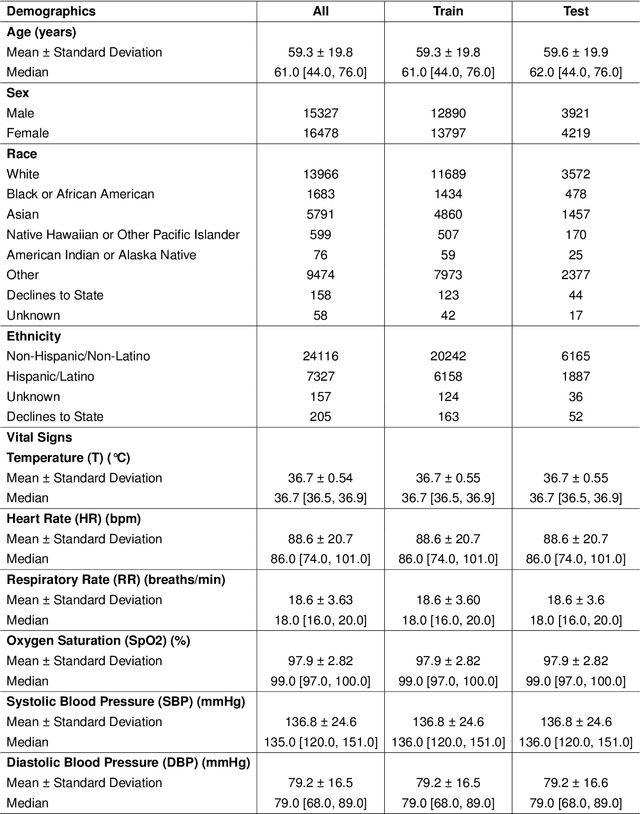
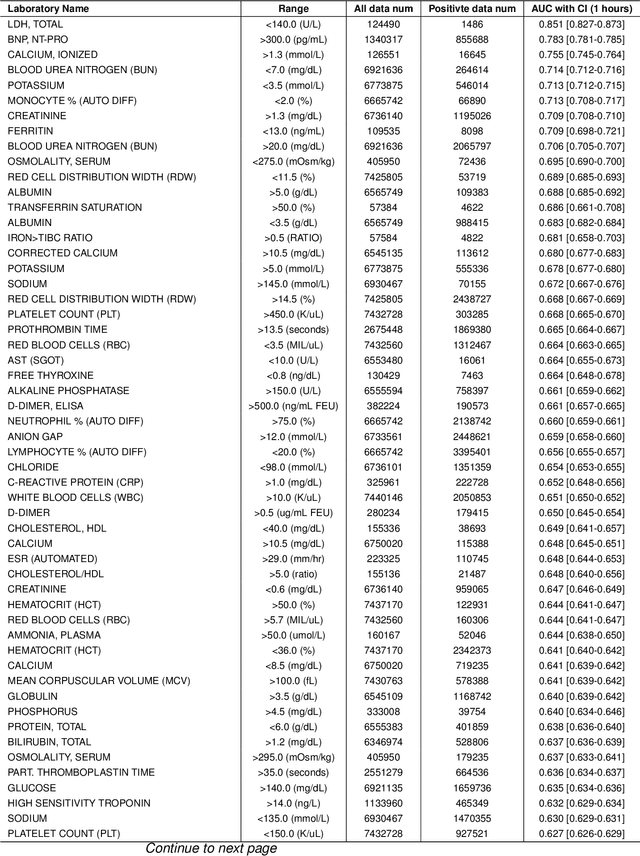
Abstract:Timely access to laboratory values is critical for clinical decision-making, yet current approaches rely on invasive venous sampling and are intrinsically delayed. Electrocardiography (ECG), as a non-invasive and widely available signal, offers a promising modality for rapid laboratory estimation. Recent progress in deep learning has enabled the extraction of latent hematological signatures from ECGs. However, existing models are constrained by low signal-to-noise ratios, substantial inter-individual variability, limited data diversity, and suboptimal generalization, especially when adapted to low-lead wearable devices. In this work, we conduct an exploratory study leveraging transfer learning to fine-tune ECGFounder, a large-scale pre-trained ECG foundation model, on the Multimodal Clinical Monitoring in the Emergency Department (MC-MED) dataset from Stanford. We generated a corpus of more than 20 million standardized ten-second ECG segments to enhance sensitivity to subtle biochemical correlates. On internal validation, the model demonstrated strong predictive performance (area under the curve above 0.65) for thirty-three laboratory indicators, moderate performance (between 0.55 and 0.65) for fifty-nine indicators, and limited performance (below 0.55) for sixteen indicators. This study provides an efficient artificial-intelligence driven solution and establishes the feasibility scope for real-time, non-invasive estimation of laboratory values.
Copy-Paste to Mitigate Large Language Model Hallucinations
Oct 01, 2025Abstract:While Retrieval-Augmented Generation (RAG) enables large language models (LLMs) to generate contextually grounded responses, contextual faithfulness remains challenging as LLMs may not consistently trust provided context, leading to hallucinations that undermine reliability. We observe an inverse correlation between response copying degree and context-unfaithful hallucinations on RAGTruth, suggesting that higher copying degrees reduce hallucinations by fostering genuine contextual belief. We propose CopyPasteLLM, obtained through two-stage high-copying response preference training. We design three prompting methods to enhance copying degree, demonstrating that high-copying responses achieve superior contextual faithfulness and hallucination control. These approaches enable a fully automated pipeline that transforms generated responses into high-copying preference data for training CopyPasteLLM. On FaithEval, ConFiQA and PubMedQA, CopyPasteLLM achieves best performance in both counterfactual and original contexts, remarkably with 12.2% to 24.5% accuracy improvements on FaithEval over the best baseline, while requiring only 365 training samples -- 1/50th of baseline data. To elucidate CopyPasteLLM's effectiveness, we propose the Context-Parameter Copying Capturing algorithm. Interestingly, this reveals that CopyPasteLLM recalibrates reliance on internal parametric knowledge rather than external knowledge during generation. All codes are available at https://github.com/longyongchao/CopyPasteLLM
Simultaneous Polysomnography and Cardiotocography Reveal Temporal Correlation Between Maternal Obstructive Sleep Apnea and Fetal Hypoxia
Apr 17, 2025Abstract:Background: Obstructive sleep apnea syndrome (OSAS) during pregnancy is common and can negatively affect fetal outcomes. However, studies on the immediate effects of maternal hypoxia on fetal heart rate (FHR) changes are lacking. Methods: We used time-synchronized polysomnography (PSG) and cardiotocography (CTG) data from two cohorts to analyze the correlation between maternal hypoxia and FHR changes (accelerations or decelerations). Maternal hypoxic event characteristics were analyzed using generalized linear modeling (GLM) to assess their associations with different FHR changes. Results: A total of 118 pregnant women participated. FHR changes were significantly associated with maternal hypoxia, primarily characterized by accelerations. A longer hypoxic duration correlated with more significant FHR accelerations (P < 0.05), while prolonged hypoxia and greater SpO2 drop were linked to FHR decelerations (P < 0.05). Both cohorts showed a transient increase in FHR during maternal hypoxia, which returned to baseline after the event resolved. Conclusion: Maternal hypoxia significantly affects FHR, suggesting that maternal OSAS may contribute to fetal hypoxia. These findings highlight the importance of maternal-fetal interactions and provide insights for future interventions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge