Gari Clifford
CARE-PD: A Multi-Site Anonymized Clinical Dataset for Parkinson's Disease Gait Assessment
Oct 05, 2025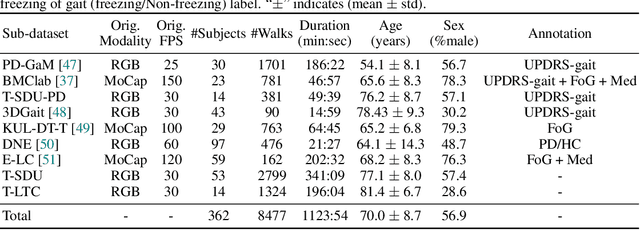
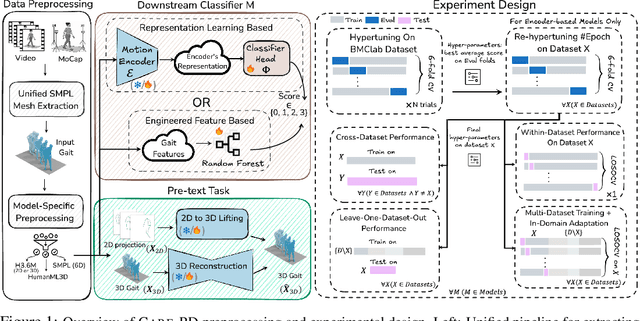
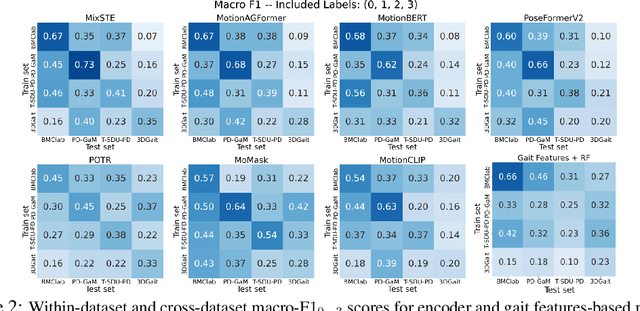
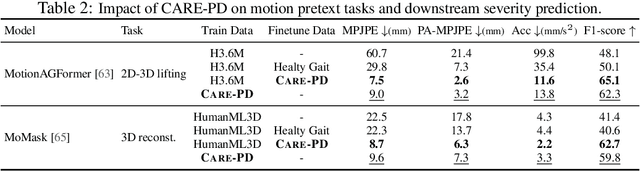
Abstract:Objective gait assessment in Parkinson's Disease (PD) is limited by the absence of large, diverse, and clinically annotated motion datasets. We introduce CARE-PD, the largest publicly available archive of 3D mesh gait data for PD, and the first multi-site collection spanning 9 cohorts from 8 clinical centers. All recordings (RGB video or motion capture) are converted into anonymized SMPL meshes via a harmonized preprocessing pipeline. CARE-PD supports two key benchmarks: supervised clinical score prediction (estimating Unified Parkinson's Disease Rating Scale, UPDRS, gait scores) and unsupervised motion pretext tasks (2D-to-3D keypoint lifting and full-body 3D reconstruction). Clinical prediction is evaluated under four generalization protocols: within-dataset, cross-dataset, leave-one-dataset-out, and multi-dataset in-domain adaptation. To assess clinical relevance, we compare state-of-the-art motion encoders with a traditional gait-feature baseline, finding that encoders consistently outperform handcrafted features. Pretraining on CARE-PD reduces MPJPE (from 60.8mm to 7.5mm) and boosts PD severity macro-F1 by 17 percentage points, underscoring the value of clinically curated, diverse training data. CARE-PD and all benchmark code are released for non-commercial research at https://neurips2025.care-pd.ca/.
From Hospital to Portables: A Universal ECG Foundation Model Built on 10+ Million Diverse Recordings
Oct 05, 2024Abstract:Artificial Intelligence (AI) has shown great promise in electrocardiogram (ECG) analysis and cardiovascular disease detection. However, developing a general AI-ECG model has been challenging due to inter-individual variability and the diversity of ECG diagnoses, limiting existing models to specific diagnostic tasks and datasets. Moreover, current AI-ECG models struggle to achieve comparable performance between single-lead and 12-lead ECGs, limiting the application of AI-ECG to portable and wearable ECG devices. To address these limitations, we introduce an ECG Foundation Model (ECGFounder), a general-purpose model that leverages real-world ECG annotations from cardiology experts to broaden the diagnostic capabilities of ECG analysis. ECGFounder is trained on over 10 million ECGs with 150 label categories from the Harvard-Emory ECG Database, enabling comprehensive cardiovascular disease diagnosis through ECG analysis. The model is designed to be both effective out-of-the-box and fine-tunable for downstream tasks, maximizing usability. More importantly, we extend its application to single-lead ECGs, enabling complex condition diagnoses and supporting various downstream tasks in mobile and remote monitoring scenarios. Experimental results demonstrate that ECGFounder achieves expert-level performance on internal validation sets for both 12-lead and single-lead ECGs, while also exhibiting strong classification performance and generalization across various diagnoses on external validation sets. When fine-tuned, ECGFounder outperforms baseline models in demographics detection, clinical event detection, and cross-modality cardiac rhythm diagnosis. The trained model and data will be publicly released upon publication through the bdsp.io. Our code is available at https://github.com/bdsp-core/ECGFounder.
ProductGraphSleepNet: Sleep Staging using Product Spatio-Temporal Graph Learning with Attentive Temporal Aggregation
Dec 09, 2022



Abstract:The classification of sleep stages plays a crucial role in understanding and diagnosing sleep pathophysiology. Sleep stage scoring relies heavily on visual inspection by an expert that is time consuming and subjective procedure. Recently, deep learning neural network approaches have been leveraged to develop a generalized automated sleep staging and account for shifts in distributions that may be caused by inherent inter/intra-subject variability, heterogeneity across datasets, and different recording environments. However, these networks ignore the connections among brain regions, and disregard the sequential connections between temporally adjacent sleep epochs. To address these issues, this work proposes an adaptive product graph learning-based graph convolutional network, named ProductGraphSleepNet, for learning joint spatio-temporal graphs along with a bidirectional gated recurrent unit and a modified graph attention network to capture the attentive dynamics of sleep stage transitions. Evaluation on two public databases: the Montreal Archive of Sleep Studies (MASS) SS3; and the SleepEDF, which contain full night polysomnography recordings of 62 and 20 healthy subjects, respectively, demonstrates performance comparable to the state-of-the-art (Accuracy: 0.867;0.838, F1-score: 0.818;0.774 and Kappa: 0.802;0.775, on each database respectively). More importantly, the proposed network makes it possible for clinicians to comprehend and interpret the learned connectivity graphs for sleep stages.
Cluster consistency: Simple yet effect robust learning algorithm on large-scale photoplethysmography for atrial fibrillation detection in the presence of real-world label noise
Nov 07, 2022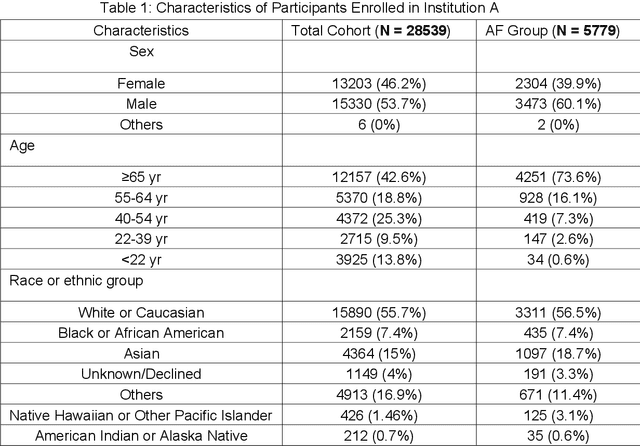
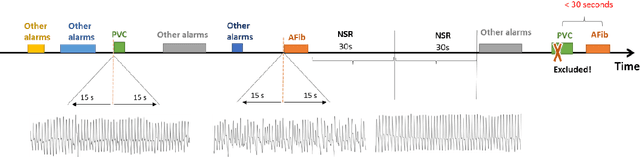
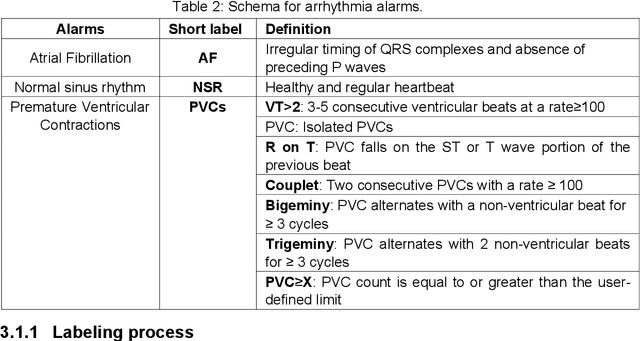
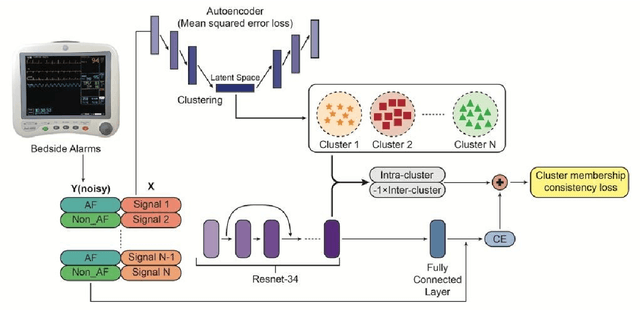
Abstract:Obtaining large-scale well-annotated is always a daunting challenge, especially in the medical research domain because of the shortage of domain expert. Instead of human annotation, in this work, we use the alarm information generated from bed-side monitor to get the pseudo label for the co-current photoplethysmography (PPG) signal. Based on this strategy, we end up with over 8 million 30-second PPG segment. To solve the label noise caused by false alarms, we propose the cluster consistency, which use an unsupervised auto-encoder (hence not subject to label noise) approach to cluster training samples into a finite number of clusters. Then the learned cluster membership is used in the subsequent supervised learning phase to force the distance in the latent space of samples in the same cluster to be small while that of samples in different clusters to be big. In the experiment, we compare with the state-of-the-art algorithms and test on external datasets. The results show the superiority of our method in both classification performance and efficiency.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge