Che Liu
CogDoc: Towards Unified thinking in Documents
Dec 14, 2025Abstract:Current document reasoning paradigms are constrained by a fundamental trade-off between scalability (processing long-context documents) and fidelity (capturing fine-grained, multimodal details). To bridge this gap, we propose CogDoc, a unified coarse-to-fine thinking framework that mimics human cognitive processes: a low-resolution "Fast Reading" phase for scalable information localization,followed by a high-resolution "Focused Thinking" phase for deep reasoning. We conduct a rigorous investigation into post-training strategies for the unified thinking framework, demonstrating that a Direct Reinforcement Learning (RL) approach outperforms RL with Supervised Fine-Tuning (SFT) initialization. Specifically, we find that direct RL avoids the "policy conflict" observed in SFT. Empirically, our 7B model achieves state-of-the-art performance within its parameter class, notably surpassing significantly larger proprietary models (e.g., GPT-4o) on challenging, visually rich document benchmarks.
Emergent Hierarchical Reasoning in LLMs through Reinforcement Learning
Sep 03, 2025



Abstract:Reinforcement Learning (RL) has proven highly effective at enhancing the complex reasoning abilities of Large Language Models (LLMs), yet underlying mechanisms driving this success remain largely opaque. Our analysis reveals that puzzling phenomena like ``aha moments", ``length-scaling'' and entropy dynamics are not disparate occurrences but hallmarks of an emergent reasoning hierarchy, akin to the separation of high-level strategic planning from low-level procedural execution in human cognition. We uncover a compelling two-phase dynamic: initially, a model is constrained by procedural correctness and must improve its low-level skills. The learning bottleneck then decisively shifts, with performance gains being driven by the exploration and mastery of high-level strategic planning. This insight exposes a core inefficiency in prevailing RL algorithms like GRPO, which apply optimization pressure agnostically and dilute the learning signal across all tokens. To address this, we propose HIerarchy-Aware Credit Assignment (HICRA), an algorithm that concentrates optimization efforts on high-impact planning tokens. HICRA significantly outperforms strong baselines, demonstrating that focusing on this strategic bottleneck is key to unlocking advanced reasoning. Furthermore, we validate semantic entropy as a superior compass for measuring strategic exploration over misleading metrics such as token-level entropy.
Knowledge to Sight: Reasoning over Visual Attributes via Knowledge Decomposition for Abnormality Grounding
Aug 06, 2025Abstract:In this work, we address the problem of grounding abnormalities in medical images, where the goal is to localize clinical findings based on textual descriptions. While generalist Vision-Language Models (VLMs) excel in natural grounding tasks, they often struggle in the medical domain due to rare, compositional, and domain-specific terms that are poorly aligned with visual patterns. Specialized medical VLMs address this challenge via large-scale domain pretraining, but at the cost of substantial annotation and computational resources. To overcome these limitations, we propose \textbf{Knowledge to Sight (K2Sight)}, a framework that introduces structured semantic supervision by decomposing clinical concepts into interpretable visual attributes, such as shape, density, and anatomical location. These attributes are distilled from domain ontologies and encoded into concise instruction-style prompts, which guide region-text alignment during training. Unlike conventional report-level supervision, our approach explicitly bridges domain knowledge and spatial structure, enabling data-efficient training of compact models. We train compact models with 0.23B and 2B parameters using only 1.5\% of the data required by state-of-the-art medical VLMs. Despite their small size and limited training data, these models achieve performance on par with or better than 7B+ medical VLMs, with up to 9.82\% improvement in $mAP_{50}$. Code and models: \href{https://lijunrio.github.io/K2Sight/}{\textcolor{SOTAPink}{https://lijunrio.github.io/K2Sight/}}.
Step-Audio 2 Technical Report
Jul 24, 2025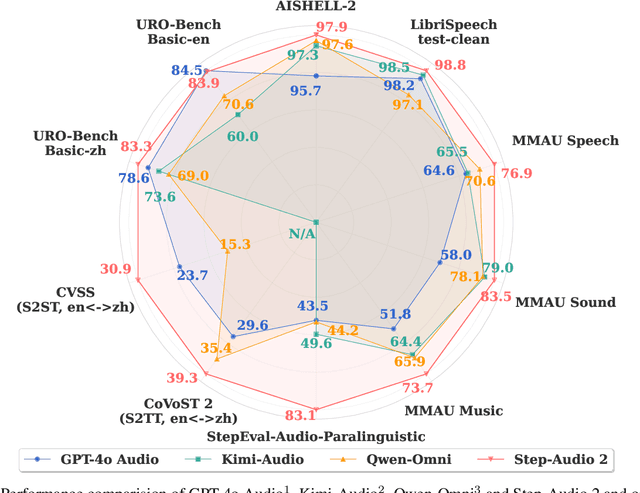

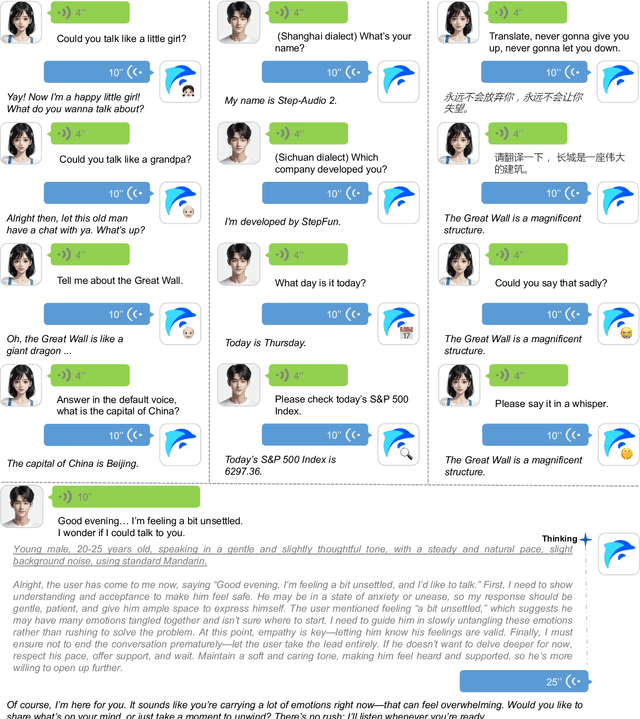

Abstract:This paper presents Step-Audio 2, an end-to-end multi-modal large language model designed for industry-strength audio understanding and speech conversation. By integrating a latent audio encoder and reasoning-centric reinforcement learning (RL), Step-Audio 2 achieves promising performance in automatic speech recognition (ASR) and audio understanding. To facilitate genuine end-to-end speech conversation, Step-Audio 2 incorporates the generation of discrete audio tokens into language modeling, significantly enhancing its responsiveness to paralinguistic information such as speaking styles and emotions. To effectively leverage the rich textual and acoustic knowledge in real-world data, Step-Audio 2 integrates retrieval-augmented generation (RAG) and is able to call external tools such as web search to mitigate hallucination and audio search to switch timbres. Trained on millions of hours of speech and audio data, Step-Audio 2 delivers intelligence and expressiveness across diverse conversational scenarios. Evaluation results demonstrate that Step-Audio 2 achieves state-of-the-art performance on various audio understanding and conversational benchmarks compared to other open-source and commercial solutions. Please visit https://github.com/stepfun-ai/Step-Audio2 for more information.
Dual form Complementary Masking for Domain-Adaptive Image Segmentation
Jul 16, 2025Abstract:Recent works have correlated Masked Image Modeling (MIM) with consistency regularization in Unsupervised Domain Adaptation (UDA). However, they merely treat masking as a special form of deformation on the input images and neglect the theoretical analysis, which leads to a superficial understanding of masked reconstruction and insufficient exploitation of its potential in enhancing feature extraction and representation learning. In this paper, we reframe masked reconstruction as a sparse signal reconstruction problem and theoretically prove that the dual form of complementary masks possesses superior capabilities in extracting domain-agnostic image features. Based on this compelling insight, we propose MaskTwins, a simple yet effective UDA framework that integrates masked reconstruction directly into the main training pipeline. MaskTwins uncovers intrinsic structural patterns that persist across disparate domains by enforcing consistency between predictions of images masked in complementary ways, enabling domain generalization in an end-to-end manner. Extensive experiments verify the superiority of MaskTwins over baseline methods in natural and biological image segmentation. These results demonstrate the significant advantages of MaskTwins in extracting domain-invariant features without the need for separate pre-training, offering a new paradigm for domain-adaptive segmentation.
BOTM: Echocardiography Segmentation via Bi-directional Optimal Token Matching
May 23, 2025Abstract:Existed echocardiography segmentation methods often suffer from anatomical inconsistency challenge caused by shape variation, partial observation and region ambiguity with similar intensity across 2D echocardiographic sequences, resulting in false positive segmentation with anatomical defeated structures in challenging low signal-to-noise ratio conditions. To provide a strong anatomical guarantee across different echocardiographic frames, we propose a novel segmentation framework named BOTM (Bi-directional Optimal Token Matching) that performs echocardiography segmentation and optimal anatomy transportation simultaneously. Given paired echocardiographic images, BOTM learns to match two sets of discrete image tokens by finding optimal correspondences from a novel anatomical transportation perspective. We further extend the token matching into a bi-directional cross-transport attention proxy to regulate the preserved anatomical consistency within the cardiac cyclic deformation in temporal domain. Extensive experimental results show that BOTM can generate stable and accurate segmentation outcomes (e.g. -1.917 HD on CAMUS2H LV, +1.9% Dice on TED), and provide a better matching interpretation with anatomical consistency guarantee.
Beyond Distillation: Pushing the Limits of Medical LLM Reasoning with Minimalist Rule-Based RL
May 23, 2025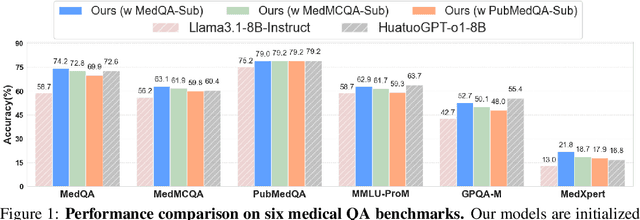
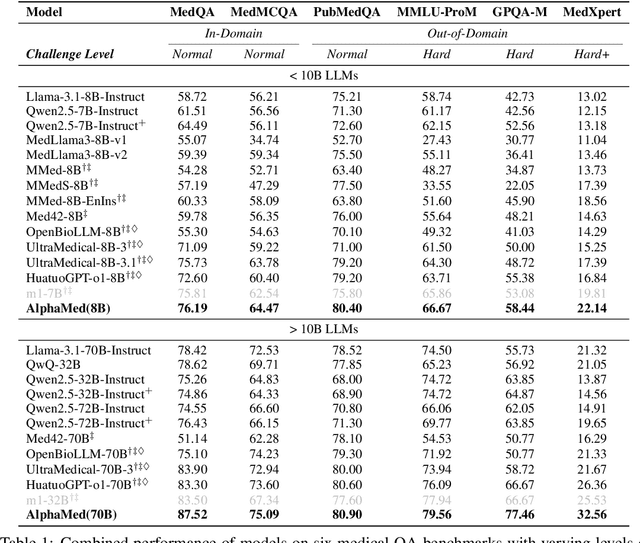
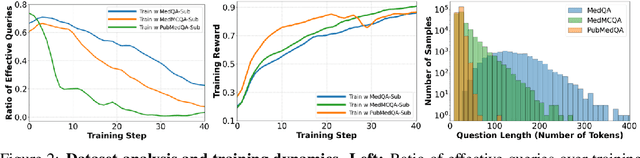
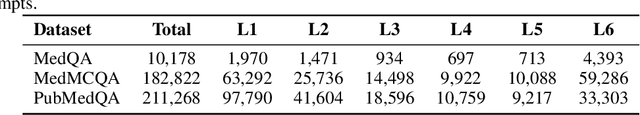
Abstract:Improving performance on complex tasks and enabling interpretable decision making in large language models (LLMs), especially for clinical applications, requires effective reasoning. Yet this remains challenging without supervised fine-tuning (SFT) on costly chain-of-thought (CoT) data distilled from closed-source models (e.g., GPT-4o). In this work, we present AlphaMed, the first medical LLM to show that reasoning capability can emerge purely through reinforcement learning (RL), using minimalist rule-based rewards on public multiple-choice QA datasets, without relying on SFT or distilled CoT data. AlphaMed achieves state-of-the-art results on six medical QA benchmarks, outperforming models trained with conventional SFT+RL pipelines. On challenging benchmarks (e.g., MedXpert), AlphaMed even surpasses larger or closed-source models such as DeepSeek-V3-671B and Claude-3.5-Sonnet. To understand the factors behind this success, we conduct a comprehensive data-centric analysis guided by three questions: (i) Can minimalist rule-based RL incentivize reasoning without distilled CoT supervision? (ii) How do dataset quantity and diversity impact reasoning? (iii) How does question difficulty shape the emergence and generalization of reasoning? Our findings show that dataset informativeness is a key driver of reasoning performance, and that minimalist RL on informative, multiple-choice QA data is effective at inducing reasoning without CoT supervision. We also observe divergent trends across benchmarks, underscoring limitations in current evaluation and the need for more challenging, reasoning-oriented medical QA benchmarks.
NOVA: A Benchmark for Anomaly Localization and Clinical Reasoning in Brain MRI
May 20, 2025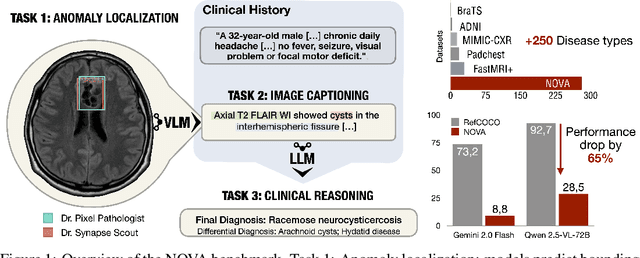

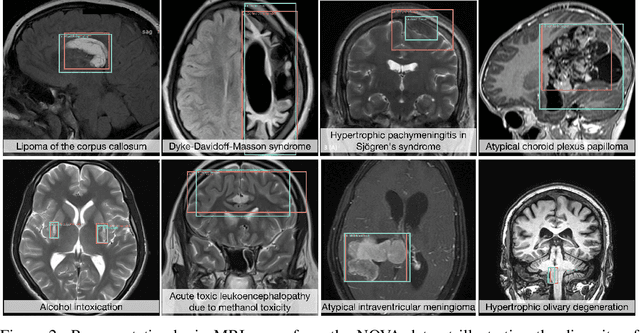
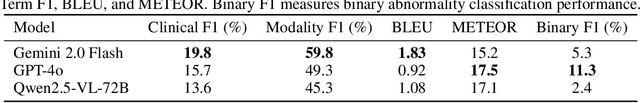
Abstract:In many real-world applications, deployed models encounter inputs that differ from the data seen during training. Out-of-distribution detection identifies whether an input stems from an unseen distribution, while open-world recognition flags such inputs to ensure the system remains robust as ever-emerging, previously $unknown$ categories appear and must be addressed without retraining. Foundation and vision-language models are pre-trained on large and diverse datasets with the expectation of broad generalization across domains, including medical imaging. However, benchmarking these models on test sets with only a few common outlier types silently collapses the evaluation back to a closed-set problem, masking failures on rare or truly novel conditions encountered in clinical use. We therefore present $NOVA$, a challenging, real-life $evaluation-only$ benchmark of $\sim$900 brain MRI scans that span 281 rare pathologies and heterogeneous acquisition protocols. Each case includes rich clinical narratives and double-blinded expert bounding-box annotations. Together, these enable joint assessment of anomaly localisation, visual captioning, and diagnostic reasoning. Because NOVA is never used for training, it serves as an $extreme$ stress-test of out-of-distribution generalisation: models must bridge a distribution gap both in sample appearance and in semantic space. Baseline results with leading vision-language models (GPT-4o, Gemini 2.0 Flash, and Qwen2.5-VL-72B) reveal substantial performance drops across all tasks, establishing NOVA as a rigorous testbed for advancing models that can detect, localize, and reason about truly unknown anomalies.
Fine-Grained ECG-Text Contrastive Learning via Waveform Understanding Enhancement
May 17, 2025Abstract:Electrocardiograms (ECGs) are essential for diagnosing cardiovascular diseases. While previous ECG-text contrastive learning methods have shown promising results, they often overlook the incompleteness of the reports. Given an ECG, the report is generated by first identifying key waveform features and then inferring the final diagnosis through these features. Despite their importance, these waveform features are often not recorded in the report as intermediate results. Aligning ECGs with such incomplete reports impedes the model's ability to capture the ECG's waveform features and limits its understanding of diagnostic reasoning based on those features. To address this, we propose FG-CLEP (Fine-Grained Contrastive Language ECG Pre-training), which aims to recover these waveform features from incomplete reports with the help of large language models (LLMs), under the challenges of hallucinations and the non-bijective relationship between waveform features and diagnoses. Additionally, considering the frequent false negatives due to the prevalence of common diagnoses in ECGs, we introduce a semantic similarity matrix to guide contrastive learning. Furthermore, we adopt a sigmoid-based loss function to accommodate the multi-label nature of ECG-related tasks. Experiments on six datasets demonstrate that FG-CLEP outperforms state-of-the-art methods in both zero-shot prediction and linear probing across these datasets.
Towards Cardiac MRI Foundation Models: Comprehensive Visual-Tabular Representations for Whole-Heart Assessment and Beyond
Apr 18, 2025Abstract:Cardiac magnetic resonance imaging is the gold standard for non-invasive cardiac assessment, offering rich spatio-temporal views of the cardiac anatomy and physiology. Patient-level health factors, such as demographics, metabolic, and lifestyle, are known to substantially influence cardiovascular health and disease risk, yet remain uncaptured by CMR alone. To holistically understand cardiac health and to enable the best possible interpretation of an individual's disease risk, CMR and patient-level factors must be jointly exploited within an integrated framework. Recent multi-modal approaches have begun to bridge this gap, yet they often rely on limited spatio-temporal data and focus on isolated clinical tasks, thereby hindering the development of a comprehensive representation for cardiac health evaluation. To overcome these limitations, we introduce ViTa, a step toward foundation models that delivers a comprehensive representation of the heart and a precise interpretation of individual disease risk. Leveraging data from 42,000 UK Biobank participants, ViTa integrates 3D+T cine stacks from short-axis and long-axis views, enabling a complete capture of the cardiac cycle. These imaging data are then fused with detailed tabular patient-level factors, enabling context-aware insights. This multi-modal paradigm supports a wide spectrum of downstream tasks, including cardiac phenotype and physiological feature prediction, segmentation, and classification of cardiac and metabolic diseases within a single unified framework. By learning a shared latent representation that bridges rich imaging features and patient context, ViTa moves beyond traditional, task-specific models toward a universal, patient-specific understanding of cardiac health, highlighting its potential to advance clinical utility and scalability in cardiac analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge