Jiayuan Zhu
MedVLM-R1: Incentivizing Medical Reasoning Capability of Vision-Language Models (VLMs) via Reinforcement Learning
Feb 26, 2025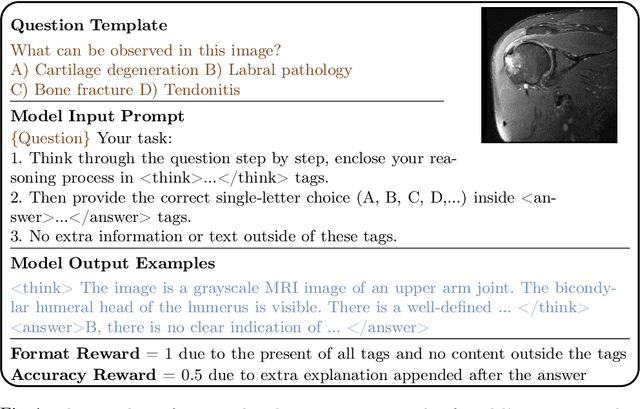
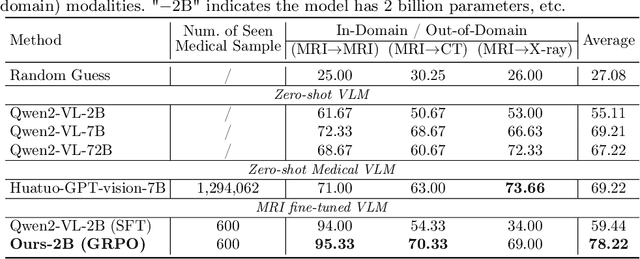
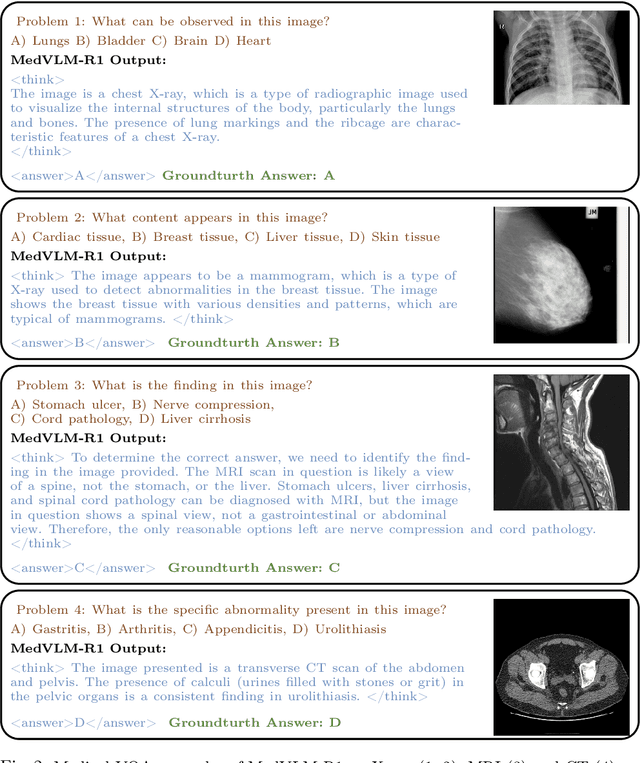
Abstract:Reasoning is a critical frontier for advancing medical image analysis, where transparency and trustworthiness play a central role in both clinician trust and regulatory approval. Although Medical Visual Language Models (VLMs) show promise for radiological tasks, most existing VLMs merely produce final answers without revealing the underlying reasoning. To address this gap, we introduce MedVLM-R1, a medical VLM that explicitly generates natural language reasoning to enhance transparency and trustworthiness. Instead of relying on supervised fine-tuning (SFT), which often suffers from overfitting to training distributions and fails to foster genuine reasoning, MedVLM-R1 employs a reinforcement learning framework that incentivizes the model to discover human-interpretable reasoning paths without using any reasoning references. Despite limited training data (600 visual question answering samples) and model parameters (2B), MedVLM-R1 boosts accuracy from 55.11% to 78.22% across MRI, CT, and X-ray benchmarks, outperforming larger models trained on over a million samples. It also demonstrates robust domain generalization under out-of-distribution tasks. By unifying medical image analysis with explicit reasoning, MedVLM-R1 marks a pivotal step toward trustworthy and interpretable AI in clinical practice.
Ask Patients with Patience: Enabling LLMs for Human-Centric Medical Dialogue with Grounded Reasoning
Feb 11, 2025



Abstract:Accurate and efficient diagnosis in online medical consultations remains a challenge for current large language models. These models often rely on single-turn interactions and lack the ability to refine their predictions through follow-up questions. Additionally, their responses frequently contain complex medical terminology, making them less accessible to non-medical users and creating barriers to effective communication. In this paper, we introduce Ask Patients with Patience (APP), the first multi-turn dialogue that enables LLMs to iteratively refine diagnoses based on grounded reasoning. By integrating medical guidelines and entropy minimization, APP improves both diagnostic accuracy and efficiency. Furthermore, it features human-centric communication that bridges the gap between user comprehension and medical terminology, significantly enhancing user accessibility and engagement. We evaluated APP using a subset of the ReMeDi dataset, comparing it with single-turn and traditional multi-turn LLM baselines. APP achieved higher similarity scores in diagnosis predictions, demonstrating better alignment with ground truth diagnoses. Entropy analysis showed that APP reduces diagnostic uncertainty more rapidly across iterations, increasing confidence in its predictions. APP also excels in user accessibility and empathy, further bridging the gap between complex medical language and user understanding. Code will be released at: https://github.com/SuperMedIntel/AskPatients.
Agentic Reasoning: Reasoning LLMs with Tools for the Deep Research
Feb 07, 2025



Abstract:We introduce Agentic Reasoning, a framework that enhances large language model (LLM) reasoning by integrating external tool-using agents. Unlike conventional LLM-based reasoning approaches, which rely solely on internal inference, Agentic Reasoning dynamically engages web search, code execution, and structured reasoning-context memory to solve complex problems requiring deep research and multi-step logical deduction. Our framework introduces the Mind Map agent, which constructs a structured knowledge graph to track logical relationships, improving deductive reasoning. Additionally, the integration of web-search and coding agents enables real-time retrieval and computational analysis, enhancing reasoning accuracy and decision-making. Evaluations on PhD-level scientific reasoning (GPQA) and domain-specific deep research tasks demonstrate that our approach significantly outperforms existing models, including leading retrieval-augmented generation (RAG) systems and closed-source LLMs. Moreover, our results indicate that agentic reasoning improves expert-level knowledge synthesis, test-time scalability, and structured problem-solving. The code is at: https://github.com/theworldofagents/Agentic-Reasoning.
SPA: Efficient User-Preference Alignment against Uncertainty in Medical Image Segmentation
Nov 23, 2024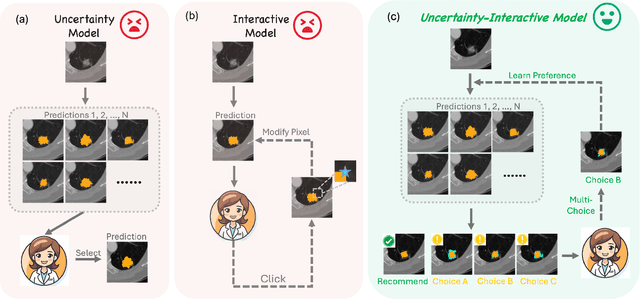
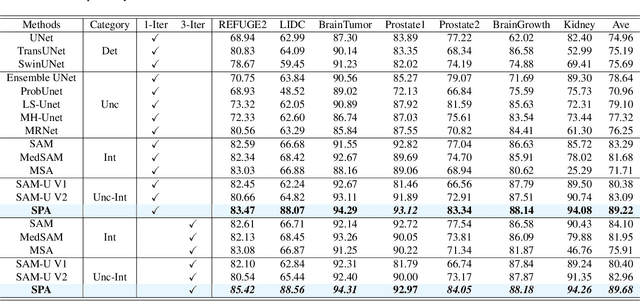
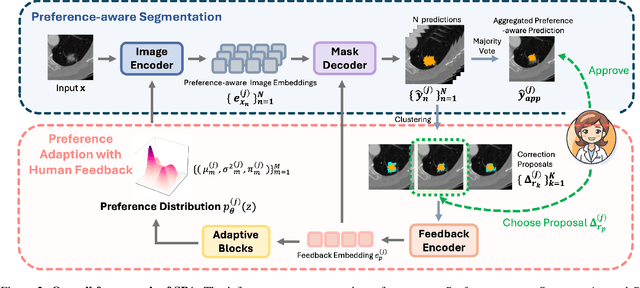
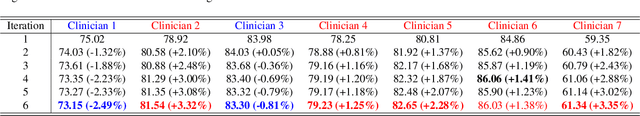
Abstract:Medical image segmentation data inherently contain uncertainty, often stemming from both imperfect image quality and variability in labeling preferences on ambiguous pixels, which depend on annotators' expertise and the clinical context of the annotations. For instance, a boundary pixel might be labeled as tumor in diagnosis to avoid under-assessment of severity, but as normal tissue in radiotherapy to prevent damage to sensitive structures. As segmentation preferences vary across downstream applications, it is often desirable for an image segmentation model to offer user-adaptable predictions rather than a fixed output. While prior uncertainty-aware and interactive methods offer adaptability, they are inefficient at test time: uncertainty-aware models require users to choose from numerous similar outputs, while interactive models demand significant user input through click or box prompts to refine segmentation. To address these challenges, we propose \textbf{SPA}, a segmentation framework that efficiently adapts to diverse test-time preferences with minimal human interaction. By presenting users a select few, distinct segmentation candidates that best capture uncertainties, it reduces clinician workload in reaching the preferred segmentation. To accommodate user preference, we introduce a probabilistic mechanism that leverages user feedback to adapt model's segmentation preference. The proposed framework is evaluated on a diverse range of medical image segmentation tasks: color fundus images, CT, and MRI. It demonstrates 1) a significant reduction in clinician time and effort compared with existing interactive segmentation approaches, 2) strong adaptability based on human feedback, and 3) state-of-the-art image segmentation performance across diverse modalities and semantic labels.
Medical Graph RAG: Towards Safe Medical Large Language Model via Graph Retrieval-Augmented Generation
Aug 08, 2024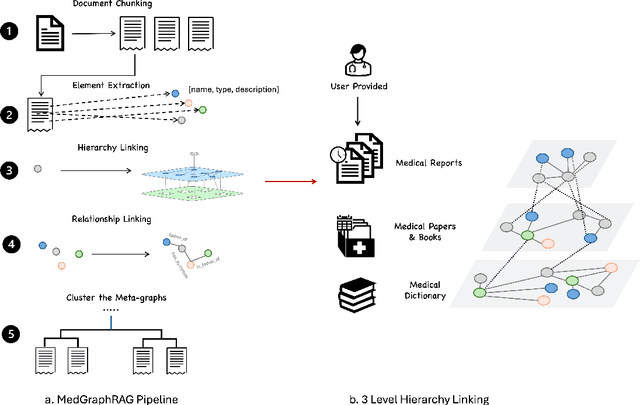
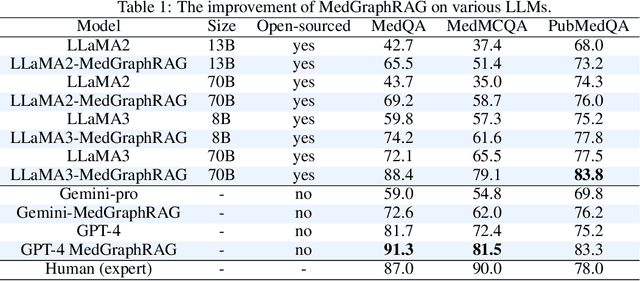
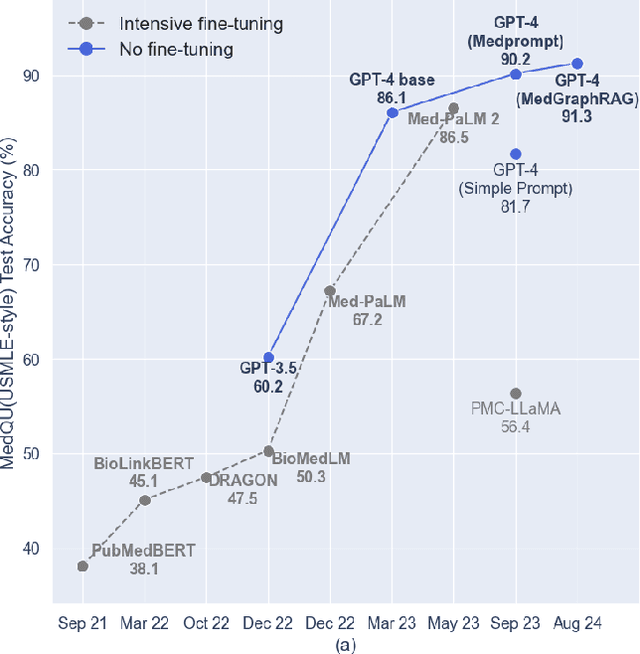
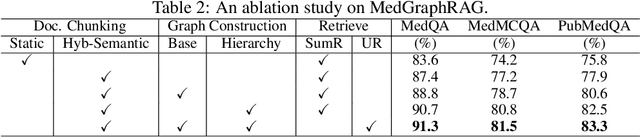
Abstract:We introduce a novel graph-based Retrieval-Augmented Generation (RAG) framework specifically designed for the medical domain, called \textbf{MedGraphRAG}, aimed at enhancing Large Language Model (LLM) capabilities and generating evidence-based results, thereby improving safety and reliability when handling private medical data. Our comprehensive pipeline begins with a hybrid static-semantic approach to document chunking, significantly improving context capture over traditional methods. Extracted entities are used to create a three-tier hierarchical graph structure, linking entities to foundational medical knowledge sourced from medical papers and dictionaries. These entities are then interconnected to form meta-graphs, which are merged based on semantic similarities to develop a comprehensive global graph. This structure supports precise information retrieval and response generation. The retrieval process employs a U-retrieve method to balance global awareness and indexing efficiency of the LLM. Our approach is validated through a comprehensive ablation study comparing various methods for document chunking, graph construction, and information retrieval. The results not only demonstrate that our hierarchical graph construction method consistently outperforms state-of-the-art models on multiple medical Q\&A benchmarks, but also confirms that the responses generated include source documentation, significantly enhancing the reliability of medical LLMs in practical applications. Code will be at: https://github.com/MedicineToken/Medical-Graph-RAG/tree/main
MedUHIP: Towards Human-In-the-Loop Medical Segmentation
Aug 03, 2024



Abstract:Although segmenting natural images has shown impressive performance, these techniques cannot be directly applied to medical image segmentation. Medical image segmentation is particularly complicated by inherent uncertainties. For instance, the ambiguous boundaries of tissues can lead to diverse but plausible annotations from different clinicians. These uncertainties cause significant discrepancies in clinical interpretations and impact subsequent medical interventions. Therefore, achieving quantitative segmentations from uncertain medical images becomes crucial in clinical practice. To address this, we propose a novel approach that integrates an \textbf{uncertainty-aware model} with \textbf{human-in-the-loop interaction}. The uncertainty-aware model proposes several plausible segmentations to address the uncertainties inherent in medical images, while the human-in-the-loop interaction iteratively modifies the segmentation under clinician supervision. This collaborative model ensures that segmentation is not solely dependent on automated techniques but is also refined through clinician expertise. As a result, our approach represents a significant advancement in the field which enhances the safety of medical image segmentation. It not only offers a comprehensive solution to produce quantitative segmentation from inherent uncertain medical images, but also establishes a synergistic balance between algorithmic precision and clincian knowledge. We evaluated our method on various publicly available multi-clinician annotated datasets: REFUGE2, LIDC-IDRI and QUBIQ. Our method showcases superior segmentation capabilities, outperforming a wide range of deterministic and uncertainty-aware models. We also demonstrated that our model produced significantly better results with fewer interactions compared to previous interactive models. We will release the code to foster further research in this area.
Medical SAM 2: Segment medical images as video via Segment Anything Model 2
Aug 01, 2024



Abstract:In this paper, we introduce Medical SAM 2 (MedSAM-2), an advanced segmentation model that utilizes the SAM 2 framework to address both 2D and 3D medical image segmentation tasks. By adopting the philosophy of taking medical images as videos, MedSAM-2 not only applies to 3D medical images but also unlocks new One-prompt Segmentation capability. That allows users to provide a prompt for just one or a specific image targeting an object, after which the model can autonomously segment the same type of object in all subsequent images, regardless of temporal relationships between the images. We evaluated MedSAM-2 across a variety of medical imaging modalities, including abdominal organs, optic discs, brain tumors, thyroid nodules, and skin lesions, comparing it against state-of-the-art models in both traditional and interactive segmentation settings. Our findings show that MedSAM-2 not only surpasses existing models in performance but also exhibits superior generalization across a range of medical image segmentation tasks. Our code will be released at: https://github.com/MedicineToken/Medical-SAM2
MGI: Multimodal Contrastive pre-training of Genomic and Medical Imaging
Jun 02, 2024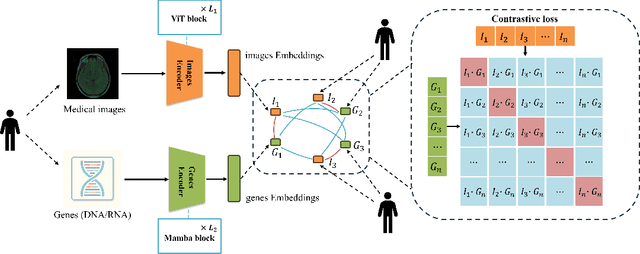
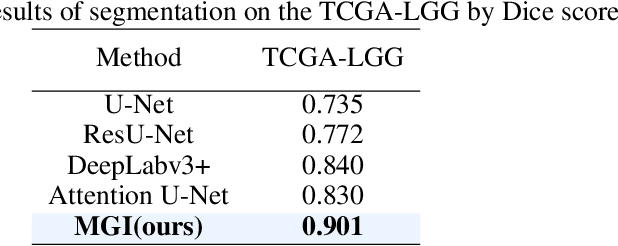
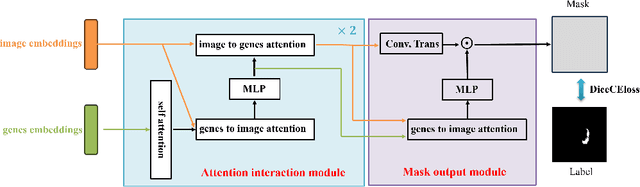
Abstract:Medicine is inherently a multimodal discipline. Medical images can reflect the pathological changes of cancer and tumors, while the expression of specific genes can influence their morphological characteristics. However, most deep learning models employed for these medical tasks are unimodal, making predictions using either image data or genomic data exclusively. In this paper, we propose a multimodal pre-training framework that jointly incorporates genomics and medical images for downstream tasks. To address the issues of high computational complexity and difficulty in capturing long-range dependencies in genes sequence modeling with MLP or Transformer architectures, we utilize Mamba to model these long genomic sequences. We aligns medical images and genes using a self-supervised contrastive learning approach which combines the Mamba as a genetic encoder and the Vision Transformer (ViT) as a medical image encoder. We pre-trained on the TCGA dataset using paired gene expression data and imaging data, and fine-tuned it for downstream tumor segmentation tasks. The results show that our model outperformed a wide range of related methods.
Not just Birds and Cars: Generic, Scalable and Explainable Models for Professional Visual Recognition
Mar 08, 2024



Abstract:Some visual recognition tasks are more challenging then the general ones as they require professional categories of images. The previous efforts, like fine-grained vision classification, primarily introduced models tailored to specific tasks, like identifying bird species or car brands with limited scalability and generalizability. This paper aims to design a scalable and explainable model to solve Professional Visual Recognition tasks from a generic standpoint. We introduce a biologically-inspired structure named Pro-NeXt and reveal that Pro-NeXt exhibits substantial generalizability across diverse professional fields such as fashion, medicine, and art-areas previously considered disparate. Our basic-sized Pro-NeXt-B surpasses all preceding task-specific models across 12 distinct datasets within 5 diverse domains. Furthermore, we find its good scaling property that scaling up Pro-NeXt in depth and width with increasing GFlops can consistently enhances its accuracy. Beyond scalability and adaptability, the intermediate features of Pro-NeXt achieve reliable object detection and segmentation performance without extra training, highlighting its solid explainability. We will release the code to foster further research in this area.
UGformer for Robust Left Atrium and Scar Segmentation Across Scanners
Oct 11, 2022
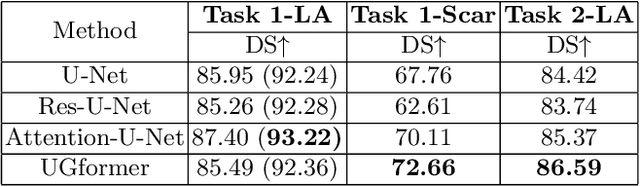
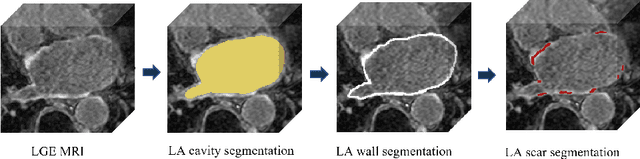
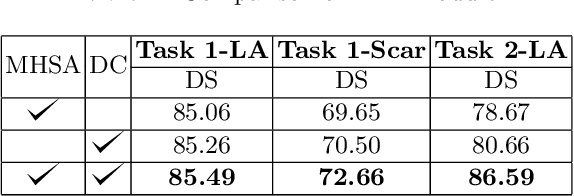
Abstract:Thanks to the capacity for long-range dependencies and robustness to irregular shapes, vision transformers and deformable convolutions are emerging as powerful vision techniques of segmentation.Meanwhile, Graph Convolution Networks (GCN) optimize local features based on global topological relationship modeling. Particularly, they have been proved to be effective in addressing issues in medical imaging segmentation tasks including multi-domain generalization for low-quality images. In this paper, we present a novel, effective, and robust framework for medical image segmentation, namely, UGformer. It unifies novel transformer blocks, GCN bridges, and convolution decoders originating from U-Net to predict left atriums (LAs) and LA scars. We have identified two appealing findings of the proposed UGformer: 1). an enhanced transformer module with deformable convolutions to improve the blending of the transformer information with convolutional information and help predict irregular LAs and scar shapes. 2). Using a bridge incorporating GCN to further overcome the difficulty of capturing condition inconsistency across different Magnetic Resonance Images scanners with various inconsistent domain information. The proposed UGformer model exhibits outstanding ability to segment the left atrium and scar on the LAScarQS 2022 dataset, outperforming several recent state-of-the-arts.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge