Konstantinos Kamnitsas
Modality-Agnostic Input Channels Enable Segmentation of Brain lesions in Multimodal MRI with Sequences Unavailable During Training
Sep 11, 2025Abstract:Segmentation models are important tools for the detection and analysis of lesions in brain MRI. Depending on the type of brain pathology that is imaged, MRI scanners can acquire multiple, different image modalities (contrasts). Most segmentation models for multimodal brain MRI are restricted to fixed modalities and cannot effectively process new ones at inference. Some models generalize to unseen modalities but may lose discriminative modality-specific information. This work aims to develop a model that can perform inference on data that contain image modalities unseen during training, previously seen modalities, and heterogeneous combinations of both, thus allowing a user to utilize any available imaging modalities. We demonstrate this is possible with a simple, thus practical alteration to the U-net architecture, by integrating a modality-agnostic input channel or pathway, alongside modality-specific input channels. To train this modality-agnostic component, we develop an image augmentation scheme that synthesizes artificial MRI modalities. Augmentations differentially alter the appearance of pathological and healthy brain tissue to create artificial contrasts between them while maintaining realistic anatomical integrity. We evaluate the method using 8 MRI databases that include 5 types of pathologies (stroke, tumours, traumatic brain injury, multiple sclerosis and white matter hyperintensities) and 8 modalities (T1, T1+contrast, T2, PD, SWI, DWI, ADC and FLAIR). The results demonstrate that the approach preserves the ability to effectively process MRI modalities encountered during training, while being able to process new, unseen modalities to improve its segmentation. Project code: https://github.com/Anthony-P-Addison/AGN-MOD-SEG
Specialised or Generic? Tokenization Choices for Radiology Language Models
Aug 13, 2025Abstract:The vocabulary used by language models (LM) - defined by the tokenizer - plays a key role in text generation quality. However, its impact remains under-explored in radiology. In this work, we address this gap by systematically comparing general, medical, and domain-specific tokenizers on the task of radiology report summarisation across three imaging modalities. We also investigate scenarios with and without LM pre-training on PubMed abstracts. Our findings demonstrate that medical and domain-specific vocabularies outperformed widely used natural language alternatives when models are trained from scratch. Pre-training partially mitigates performance differences between tokenizers, whilst the domain-specific tokenizers achieve the most favourable results. Domain-specific tokenizers also reduce memory requirements due to smaller vocabularies and shorter sequences. These results demonstrate that adapting the vocabulary of LMs to the clinical domain provides practical benefits, including improved performance and reduced computational demands, making such models more accessible and effective for both research and real-world healthcare settings.
DIsoN: Decentralized Isolation Networks for Out-of-Distribution Detection in Medical Imaging
Jun 10, 2025



Abstract:Safe deployment of machine learning (ML) models in safety-critical domains such as medical imaging requires detecting inputs with characteristics not seen during training, known as out-of-distribution (OOD) detection, to prevent unreliable predictions. Effective OOD detection after deployment could benefit from access to the training data, enabling direct comparison between test samples and the training data distribution to identify differences. State-of-the-art OOD detection methods, however, either discard training data after deployment or assume that test samples and training data are centrally stored together, an assumption that rarely holds in real-world settings. This is because shipping training data with the deployed model is usually impossible due to the size of training databases, as well as proprietary or privacy constraints. We introduce the Isolation Network, an OOD detection framework that quantifies the difficulty of separating a target test sample from the training data by solving a binary classification task. We then propose Decentralized Isolation Networks (DIsoN), which enables the comparison of training and test data when data-sharing is impossible, by exchanging only model parameters between the remote computational nodes of training and deployment. We further extend DIsoN with class-conditioning, comparing a target sample solely with training data of its predicted class. We evaluate DIsoN on four medical imaging datasets (dermatology, chest X-ray, breast ultrasound, histopathology) across 12 OOD detection tasks. DIsoN performs favorably against existing methods while respecting data-privacy. This decentralized OOD detection framework opens the way for a new type of service that ML developers could provide along with their models: providing remote, secure utilization of their training data for OOD detection services. Code will be available upon acceptance at: *****
IterMask3D: Unsupervised Anomaly Detection and Segmentation with Test-Time Iterative Mask Refinement in 3D Brain MR
Apr 07, 2025Abstract:Unsupervised anomaly detection and segmentation methods train a model to learn the training distribution as 'normal'. In the testing phase, they identify patterns that deviate from this normal distribution as 'anomalies'. To learn the `normal' distribution, prevailing methods corrupt the images and train a model to reconstruct them. During testing, the model attempts to reconstruct corrupted inputs based on the learned 'normal' distribution. Deviations from this distribution lead to high reconstruction errors, which indicate potential anomalies. However, corrupting an input image inevitably causes information loss even in normal regions, leading to suboptimal reconstruction and an increased risk of false positives. To alleviate this, we propose IterMask3D, an iterative spatial mask-refining strategy designed for 3D brain MRI. We iteratively spatially mask areas of the image as corruption and reconstruct them, then shrink the mask based on reconstruction error. This process iteratively unmasks 'normal' areas to the model, whose information further guides reconstruction of 'normal' patterns under the mask to be reconstructed accurately, reducing false positives. In addition, to achieve better reconstruction performance, we also propose using high-frequency image content as additional structural information to guide the reconstruction of the masked area. Extensive experiments on the detection of both synthetic and real-world imaging artifacts, as well as segmentation of various pathological lesions across multiple MRI sequences, consistently demonstrate the effectiveness of our proposed method.
Continuous Online Adaptation Driven by User Interaction for Medical Image Segmentation
Mar 09, 2025Abstract:Interactive segmentation models use real-time user interactions, such as mouse clicks, as extra inputs to dynamically refine the model predictions. After model deployment, user corrections of model predictions could be used to adapt the model to the post-deployment data distribution, countering distribution-shift and enhancing reliability. Motivated by this, we introduce an online adaptation framework that enables an interactive segmentation model to continuously learn from user interaction and improve its performance on new data distributions, as it processes a sequence of test images. We introduce the Gaussian Point Loss function to train the model how to leverage user clicks, along with a two-stage online optimization method that adapts the model using the corrected predictions generated via user interactions. We demonstrate that this simple and therefore practical approach is very effective. Experiments on 5 fundus and 4 brain MRI databases demonstrate that our method outperforms existing approaches under various data distribution shifts, including segmentation of image modalities and pathologies not seen during training.
FedPIA -- Permuting and Integrating Adapters leveraging Wasserstein Barycenters for Finetuning Foundation Models in Multi-Modal Federated Learning
Dec 19, 2024Abstract:Large Vision-Language Models typically require large text and image datasets for effective fine-tuning. However, collecting data from various sites, especially in healthcare, is challenging due to strict privacy regulations. An alternative is to fine-tune these models on end-user devices, such as in medical clinics, without sending data to a server. These local clients typically have limited computing power and small datasets, which are not enough for fully fine-tuning large VLMs on their own. A naive solution to these scenarios is to leverage parameter-efficient fine-tuning (PEFT) strategies and apply federated learning (FL) algorithms to combine the learned adapter weights, thereby respecting the resource limitations and data privacy. However, this approach does not fully leverage the knowledge from multiple adapters trained on diverse data distributions and for diverse tasks. The adapters are adversely impacted by data heterogeneity and task heterogeneity across clients resulting in suboptimal convergence. To this end, we propose a novel framework called FedPIA that improves upon the naive combinations of FL and PEFT by introducing Permutation and Integration of the local Adapters in the server and global Adapters in the clients exploiting Wasserstein barycenters for improved blending of client-specific and client-agnostic knowledge. This layerwise permutation helps to bridge the gap in the parameter space of local and global adapters before integration. We conduct over 2000 client-level experiments utilizing 48 medical image datasets across five different medical vision-language FL task settings encompassing visual question answering as well as image and report-based multi-label disease detection. Our experiments involving diverse client settings, ten different modalities, and two VLM backbones demonstrate that FedPIA consistently outperforms the state-of-the-art PEFT-FL baselines.
SPA: Efficient User-Preference Alignment against Uncertainty in Medical Image Segmentation
Nov 23, 2024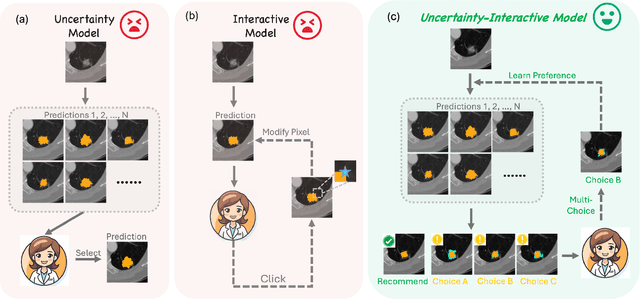
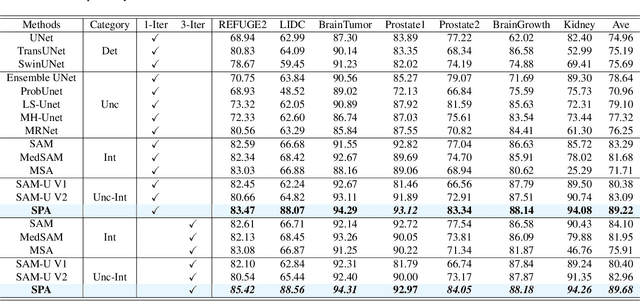
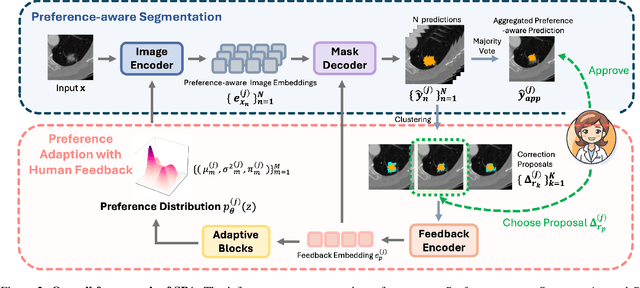
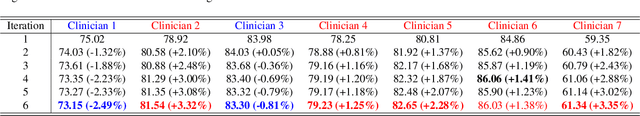
Abstract:Medical image segmentation data inherently contain uncertainty, often stemming from both imperfect image quality and variability in labeling preferences on ambiguous pixels, which depend on annotators' expertise and the clinical context of the annotations. For instance, a boundary pixel might be labeled as tumor in diagnosis to avoid under-assessment of severity, but as normal tissue in radiotherapy to prevent damage to sensitive structures. As segmentation preferences vary across downstream applications, it is often desirable for an image segmentation model to offer user-adaptable predictions rather than a fixed output. While prior uncertainty-aware and interactive methods offer adaptability, they are inefficient at test time: uncertainty-aware models require users to choose from numerous similar outputs, while interactive models demand significant user input through click or box prompts to refine segmentation. To address these challenges, we propose \textbf{SPA}, a segmentation framework that efficiently adapts to diverse test-time preferences with minimal human interaction. By presenting users a select few, distinct segmentation candidates that best capture uncertainties, it reduces clinician workload in reaching the preferred segmentation. To accommodate user preference, we introduce a probabilistic mechanism that leverages user feedback to adapt model's segmentation preference. The proposed framework is evaluated on a diverse range of medical image segmentation tasks: color fundus images, CT, and MRI. It demonstrates 1) a significant reduction in clinician time and effort compared with existing interactive segmentation approaches, 2) strong adaptability based on human feedback, and 3) state-of-the-art image segmentation performance across diverse modalities and semantic labels.
F$^3$OCUS -- Federated Finetuning of Vision-Language Foundation Models with Optimal Client Layer Updating Strategy via Multi-objective Meta-Heuristics
Nov 17, 2024



Abstract:Effective training of large Vision-Language Models (VLMs) on resource-constrained client devices in Federated Learning (FL) requires the usage of parameter-efficient fine-tuning (PEFT) strategies. To this end, we demonstrate the impact of two factors \textit{viz.}, client-specific layer importance score that selects the most important VLM layers for fine-tuning and inter-client layer diversity score that encourages diverse layer selection across clients for optimal VLM layer selection. We first theoretically motivate and leverage the principal eigenvalue magnitude of layerwise Neural Tangent Kernels and show its effectiveness as client-specific layer importance score. Next, we propose a novel layer updating strategy dubbed F$^3$OCUS that jointly optimizes the layer importance and diversity factors by employing a data-free, multi-objective, meta-heuristic optimization on the server. We explore 5 different meta-heuristic algorithms and compare their effectiveness for selecting model layers and adapter layers towards PEFT-FL. Furthermore, we release a new MedVQA-FL dataset involving overall 707,962 VQA triplets and 9 modality-specific clients and utilize it to train and evaluate our method. Overall, we conduct more than 10,000 client-level experiments on 6 Vision-Language FL task settings involving 58 medical image datasets and 4 different VLM architectures of varying sizes to demonstrate the effectiveness of the proposed method.
Evaluating Reliability in Medical DNNs: A Critical Analysis of Feature and Confidence-Based OOD Detection
Aug 30, 2024Abstract:Reliable use of deep neural networks (DNNs) for medical image analysis requires methods to identify inputs that differ significantly from the training data, called out-of-distribution (OOD), to prevent erroneous predictions. OOD detection methods can be categorised as either confidence-based (using the model's output layer for OOD detection) or feature-based (not using the output layer). We created two new OOD benchmarks by dividing the D7P (dermatology) and BreastMNIST (ultrasound) datasets into subsets which either contain or don't contain an artefact (rulers or annotations respectively). Models were trained with artefact-free images, and images with the artefacts were used as OOD test sets. For each OOD image, we created a counterfactual by manually removing the artefact via image processing, to assess the artefact's impact on the model's predictions. We show that OOD artefacts can boost a model's softmax confidence in its predictions, due to correlations in training data among other factors. This contradicts the common assumption that OOD artefacts should lead to more uncertain outputs, an assumption on which most confidence-based methods rely. We use this to explain why feature-based methods (e.g. Mahalanobis score) typically have greater OOD detection performance than confidence-based methods (e.g. MCP). However, we also show that feature-based methods typically perform worse at distinguishing between inputs that lead to correct and incorrect predictions (for both OOD and ID data). Following from these insights, we argue that a combination of feature-based and confidence-based methods should be used within DNN pipelines to mitigate their respective weaknesses. These project's code and OOD benchmarks are available at: https://github.com/HarryAnthony/Evaluating_OOD_detection.
Quality Control for Radiology Report Generation Models via Auxiliary Auditing Components
Jul 31, 2024Abstract:Automation of medical image interpretation could alleviate bottlenecks in diagnostic workflows, and has become of particular interest in recent years due to advancements in natural language processing. Great strides have been made towards automated radiology report generation via AI, yet ensuring clinical accuracy in generated reports is a significant challenge, hindering deployment of such methods in clinical practice. In this work we propose a quality control framework for assessing the reliability of AI-generated radiology reports with respect to semantics of diagnostic importance using modular auxiliary auditing components (AC). Evaluating our pipeline on the MIMIC-CXR dataset, our findings show that incorporating ACs in the form of disease-classifiers can enable auditing that identifies more reliable reports, resulting in higher F1 scores compared to unfiltered generated reports. Additionally, leveraging the confidence of the AC labels further improves the audit's effectiveness.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge