Xiaomeng Li
Electronic and Computer Engineering, Hong Kong University of Science and Technology, China
Distribution-Aware Reward Estimation for Test-Time Reinforcement Learning
Jan 29, 2026Abstract:Test-time reinforcement learning (TTRL) enables large language models (LLMs) to self-improve on unlabeled inputs, but its effectiveness critically depends on how reward signals are estimated without ground-truth supervision. Most existing TTRL methods rely on majority voting (MV) over rollouts to produce deterministic rewards, implicitly assuming that the majority rollout provides a reliable learning signal. We show that this assumption is fragile: MV reduces the rollout distribution into a single outcome, discarding information about non-majority but correct actions candidates, and yields systematically biased reward estimates. To address this, we propose Distribution-AwareReward Estimation (DARE), which shifts reward estimation from a single majority outcome to the full empirical rollout distribution. DARE further augments this distribution-based reward with an exploration bonus and a distribution pruning mechanism for non-majority rollout exploration and reward denoise, yielding a more informative and robust reward estimation. Extensive experiments on challenging reasoning benchmarks show that DARE improves optimization stability and final performance over recent baselines, achieving relative improvements of 25.3% on challenging AIME 2024 and 5.3% on AMC.
Parallelism and Generation Order in Masked Diffusion Language Models: Limits Today, Potential Tomorrow
Jan 22, 2026Abstract:Masked Diffusion Language Models (MDLMs) promise parallel token generation and arbitrary-order decoding, yet it remains unclear to what extent current models truly realize these capabilities. We characterize MDLM behavior along two dimensions -- parallelism strength and generation order -- using Average Finalization Parallelism (AFP) and Kendall's tau. We evaluate eight mainstream MDLMs (up to 100B parameters) on 58 benchmarks spanning knowledge, reasoning, and programming. The results show that MDLMs still lag behind comparably sized autoregressive models, mainly because parallel probabilistic modeling weakens inter-token dependencies. Meanwhile, MDLMs exhibit adaptive decoding behavior: their parallelism and generation order vary significantly with the task domain, the stage of reasoning, and whether the output is correct. On tasks that require "backward information" (e.g., Sudoku), MDLMs adopt a solution order that tends to fill easier Sudoku blanks first, highlighting their advantages. Finally, we provide theoretical motivation and design insights supporting a Generate-then-Edit paradigm, which mitigates dependency loss while retaining the efficiency of parallel decoding.
Incentivizing Cardiologist-Like Reasoning in MLLMs for Interpretable Echocardiographic Diagnosis
Jan 13, 2026Abstract:Echocardiographic diagnosis is vital for cardiac screening yet remains challenging. Existing echocardiography foundation models do not effectively capture the relationships between quantitative measurements and clinical manifestations, whereas medical reasoning multimodal large language models (MLLMs) require costly construction of detailed reasoning paths and remain ineffective at directly incorporating such echocardiographic priors into their reasoning. To address these limitations, we propose a novel approach comprising Cardiac Reasoning Template (CRT) and CardiacMind to enhance MLLM's echocardiographic reasoning by introducing cardiologist-like mindset. Specifically, CRT provides stepwise canonical diagnostic procedures for complex cardiac diseases to streamline reasoning path construction without the need for costly case-by-case verification. To incentivize reasoning MLLM under CRT, we develop CardiacMind, a new reinforcement learning scheme with three novel rewards: Procedural Quantity Reward (PQtR), Procedural Quality Reward (PQlR), and Echocardiographic Semantic Reward (ESR). PQtR promotes detailed reasoning; PQlR promotes integration of evidence across views and modalities, while ESR grounds stepwise descriptions in visual content. Our methods show a 48% improvement in multiview echocardiographic diagnosis for 15 complex cardiac diseases and a 5% improvement on CardiacNet-PAH over prior methods. The user study on our method's reasoning outputs shows 93.33% clinician agreement with cardiologist-like reasoning logic. Our code will be available.
OFL-SAM2: Prompt SAM2 with Online Few-shot Learner for Efficient Medical Image Segmentation
Dec 31, 2025Abstract:The Segment Anything Model 2 (SAM2) has demonstrated remarkable promptable visual segmentation capabilities in video data, showing potential for extension to medical image segmentation (MIS) tasks involving 3D volumes and temporally correlated 2D image sequences. However, adapting SAM2 to MIS presents several challenges, including the need for extensive annotated medical data for fine-tuning and high-quality manual prompts, which are both labor-intensive and require intervention from medical experts. To address these challenges, we introduce OFL-SAM2, a prompt-free SAM2 framework for label-efficient MIS. Our core idea is to leverage limited annotated samples to train a lightweight mapping network that captures medical knowledge and transforms generic image features into target features, thereby providing additional discriminative target representations for each frame and eliminating the need for manual prompts. Crucially, the mapping network supports online parameter update during inference, enhancing the model's generalization across test sequences. Technically, we introduce two key components: (1) an online few-shot learner that trains the mapping network to generate target features using limited data, and (2) an adaptive fusion module that dynamically integrates the target features with the memory-attention features generated by frozen SAM2, leading to accurate and robust target representation. Extensive experiments on three diverse MIS datasets demonstrate that OFL-SAM2 achieves state-of-the-art performance with limited training data.
Boosting RL-Based Visual Reasoning with Selective Adversarial Entropy Intervention
Dec 11, 2025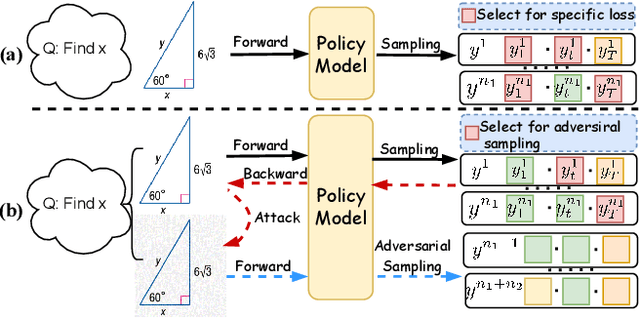
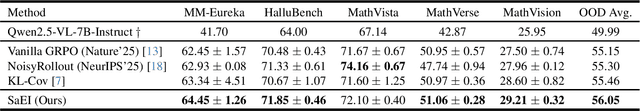
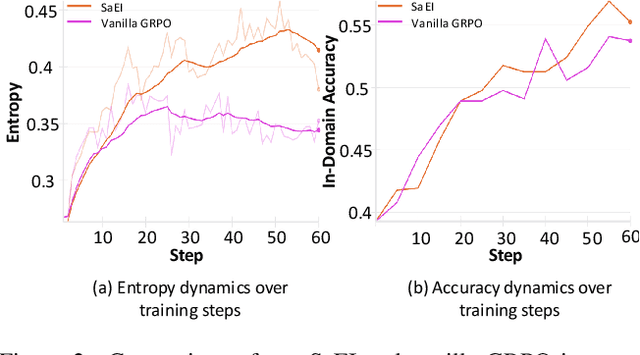
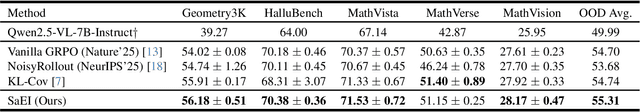
Abstract:Recently, reinforcement learning (RL) has become a common choice in enhancing the reasoning capabilities of vision-language models (VLMs). Considering existing RL-based finetuning methods, entropy intervention turns out to be an effective way to benefit exploratory ability, thereby improving policy performance. Notably, most existing studies intervene in entropy by simply controlling the update of specific tokens during policy optimization of RL. They ignore the entropy intervention during the RL sampling that can boost the performance of GRPO by improving the diversity of responses. In this paper, we propose Selective-adversarial Entropy Intervention, namely SaEI, which enhances policy entropy by distorting the visual input with the token-selective adversarial objective coming from the entropy of sampled responses. Specifically, we first propose entropy-guided adversarial sampling (EgAS) that formulates the entropy of sampled responses as an adversarial objective. Then, the corresponding adversarial gradient can be used to attack the visual input for producing adversarial samples, allowing the policy model to explore a larger answer space during RL sampling. Then, we propose token-selective entropy computation (TsEC) to maximize the effectiveness of adversarial attack in EgAS without distorting factual knowledge within VLMs. Extensive experiments on both in-domain and out-of-domain datasets show that our proposed method can greatly improve policy exploration via entropy intervention, to boost reasoning capabilities. Code will be released once the paper is accepted.
Contrastive Learning for Semi-Supervised Deep Regression with Generalized Ordinal Rankings from Spectral Seriation
Dec 10, 2025Abstract:Contrastive learning methods enforce label distance relationships in feature space to improve representation capability for regression models. However, these methods highly depend on label information to correctly recover ordinal relationships of features, limiting their applications to semi-supervised regression. In this work, we extend contrastive regression methods to allow unlabeled data to be used in the semi-supervised setting, thereby reducing the dependence on costly annotations. Particularly we construct the feature similarity matrix with both labeled and unlabeled samples in a mini-batch to reflect inter-sample relationships, and an accurate ordinal ranking of involved unlabeled samples can be recovered through spectral seriation algorithms if the level of error is within certain bounds. The introduction of labeled samples above provides regularization of the ordinal ranking with guidance from the ground-truth label information, making the ranking more reliable. To reduce feature perturbations, we further utilize the dynamic programming algorithm to select robust features for the matrix construction. The recovered ordinal relationship is then used for contrastive learning on unlabeled samples, and we thus allow more data to be used for feature representation learning, thereby achieving more robust results. The ordinal rankings can also be used to supervise predictions on unlabeled samples, serving as an additional training signal. We provide theoretical guarantees and empirical verification through experiments on various datasets, demonstrating that our method can surpass existing state-of-the-art semi-supervised deep regression methods. Our code have been released on https://github.com/xmed-lab/CLSS.
Grounded by Experience: Generative Healthcare Prediction Augmented with Hierarchical Agentic Retrieval
Nov 17, 2025Abstract:Accurate healthcare prediction is critical for improving patient outcomes and reducing operational costs. Bolstered by growing reasoning capabilities, large language models (LLMs) offer a promising path to enhance healthcare predictions by drawing on their rich parametric knowledge. However, LLMs are prone to factual inaccuracies due to limitations in the reliability and coverage of their embedded knowledge. While retrieval-augmented generation (RAG) frameworks, such as GraphRAG and its variants, have been proposed to mitigate these issues by incorporating external knowledge, they face two key challenges in the healthcare scenario: (1) identifying the clinical necessity to activate the retrieval mechanism, and (2) achieving synergy between the retriever and the generator to craft contextually appropriate retrievals. To address these challenges, we propose GHAR, a \underline{g}enerative \underline{h}ierarchical \underline{a}gentic \underline{R}AG framework that simultaneously resolves when to retrieve and how to optimize the collaboration between submodules in healthcare. Specifically, for the first challenge, we design a dual-agent architecture comprising Agent-Top and Agent-Low. Agent-Top acts as the primary physician, iteratively deciding whether to rely on parametric knowledge or to initiate retrieval, while Agent-Low acts as the consulting service, summarising all task-relevant knowledge once retrieval was triggered. To tackle the second challenge, we innovatively unify the optimization of both agents within a formal Markov Decision Process, designing diverse rewards to align their shared goal of accurate prediction while preserving their distinct roles. Extensive experiments on three benchmark datasets across three popular tasks demonstrate our superiority over state-of-the-art baselines, highlighting the potential of hierarchical agentic RAG in advancing healthcare systems.
Radiology Workflow-Guided Hierarchical Reinforcement Fine-Tuning for Medical Report Generation
Nov 13, 2025Abstract:Radiologists compose diagnostic reports through a structured workflow: they describe visual findings, summarize them into impressions, and carefully refine statements in clinically critical cases. However, most existing medical report generation (MRG) systems treat reports as flat sequences, overlooking this hierarchical organization and leading to inconsistencies between descriptive and diagnostic content. To align model behavior with real-world reporting practices, we propose RadFlow, a hierarchical workflow-guided reinforcement optimization framework that explicitly models the structured nature of clinical reporting. RadFlow introduces a clinically grounded reward hierarchy that mirrors the organization of radiological reports. At the global level, the reward integrates linguistic fluency, medical-domain correctness, and cross-sectional consistency between Finding and Impression, promoting coherent and clinically faithful narratives. At the local level, a section-specific reward emphasizes Impression quality, reflecting its central role in diagnostic accuracy. Furthermore, a critical-aware policy optimization mechanism adaptively regularizes learning for high-risk or clinically sensitive cases, emulating the cautious refinement behavior of radiologists when documenting critical findings. Together, these components translate the structured reporting paradigm into the reinforcement fine-tuning process, enabling the model to generate reports that are both linguistically consistent and clinically aligned. Experiments on chest X-ray and carotid ultrasound datasets demonstrate that RadFlow consistently improves diagnostic coherence and overall report quality compared with state-of-the-art baselines.
Evaluating LLM Understanding via Structured Tabular Decision Simulations
Nov 07, 2025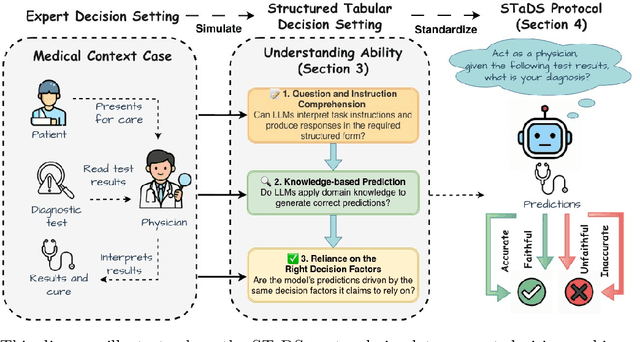
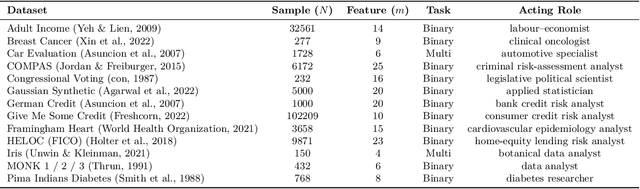
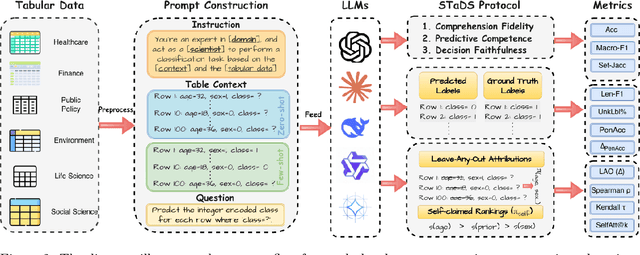
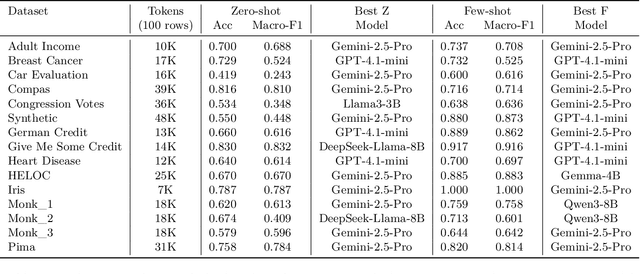
Abstract:Large language models (LLMs) often achieve impressive predictive accuracy, yet correctness alone does not imply genuine understanding. True LLM understanding, analogous to human expertise, requires making consistent, well-founded decisions across multiple instances and diverse domains, relying on relevant and domain-grounded decision factors. We introduce Structured Tabular Decision Simulations (STaDS), a suite of expert-like decision settings that evaluate LLMs as if they were professionals undertaking structured decision ``exams''. In this context, understanding is defined as the ability to identify and rely on the correct decision factors, features that determine outcomes within a domain. STaDS jointly assesses understanding through: (i) question and instruction comprehension, (ii) knowledge-based prediction, and (iii) reliance on relevant decision factors. By analyzing 9 frontier LLMs across 15 diverse decision settings, we find that (a) most models struggle to achieve consistently strong accuracy across diverse domains; (b) models can be accurate yet globally unfaithful, and there are frequent mismatches between stated rationales and factors driving predictions. Our findings highlight the need for global-level understanding evaluation protocols and advocate for novel frameworks that go beyond accuracy to enhance LLMs' understanding ability.
MedSapiens: Taking a Pose to Rethink Medical Imaging Landmark Detection
Nov 06, 2025Abstract:This paper does not introduce a novel architecture; instead, it revisits a fundamental yet overlooked baseline: adapting human-centric foundation models for anatomical landmark detection in medical imaging. While landmark detection has traditionally relied on domain-specific models, the emergence of large-scale pre-trained vision models presents new opportunities. In this study, we investigate the adaptation of Sapiens, a human-centric foundation model designed for pose estimation, to medical imaging through multi-dataset pretraining, establishing a new state of the art across multiple datasets. Our proposed model, MedSapiens, demonstrates that human-centric foundation models, inherently optimized for spatial pose localization, provide strong priors for anatomical landmark detection, yet this potential has remained largely untapped. We benchmark MedSapiens against existing state-of-the-art models, achieving up to 5.26% improvement over generalist models and up to 21.81% improvement over specialist models in the average success detection rate (SDR). To further assess MedSapiens adaptability to novel downstream tasks with few annotations, we evaluate its performance in limited-data settings, achieving 2.69% improvement over the few-shot state of the art in SDR. Code and model weights are available at https://github.com/xmed-lab/MedSapiens .
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge