Jiewen Yang
AI-Enabled Accurate Non-Invasive Assessment of Pulmonary Hypertension Progression via Multi-Modal Echocardiography
May 12, 2025Abstract:Echocardiographers can detect pulmonary hypertension using Doppler echocardiography; however, accurately assessing its progression often proves challenging. Right heart catheterization (RHC), the gold standard for precise evaluation, is invasive and unsuitable for routine use, limiting its practicality for timely diagnosis and monitoring of pulmonary hypertension progression. Here, we propose MePH, a multi-view, multi-modal vision-language model to accurately assess pulmonary hypertension progression using non-invasive echocardiography. We constructed a large dataset comprising paired standardized echocardiogram videos, spectral images and RHC data, covering 1,237 patient cases from 12 medical centers. For the first time, MePH precisely models the correlation between non-invasive multi-view, multi-modal echocardiography and the pressure and resistance obtained via RHC. We show that MePH significantly outperforms echocardiographers' assessments using echocardiography, reducing the mean absolute error in estimating mean pulmonary arterial pressure (mPAP) and pulmonary vascular resistance (PVR) by 49.73% and 43.81%, respectively. In eight independent external hospitals, MePH achieved a mean absolute error of 3.147 for PVR assessment. Furthermore, MePH achieved an area under the curve of 0.921, surpassing echocardiographers (area under the curve of 0.842) in accurately predicting the severity of pulmonary hypertension, whether mild or severe. A prospective study demonstrated that MePH can predict treatment efficacy for patients. Our work provides pulmonary hypertension patients with a non-invasive and timely method for monitoring disease progression, improving the accuracy and efficiency of pulmonary hypertension management while enabling earlier interventions and more personalized treatment decisions.
DeepSparse: A Foundation Model for Sparse-View CBCT Reconstruction
May 05, 2025Abstract:Cone-beam computed tomography (CBCT) is a critical 3D imaging technology in the medical field, while the high radiation exposure required for high-quality imaging raises significant concerns, particularly for vulnerable populations. Sparse-view reconstruction reduces radiation by using fewer X-ray projections while maintaining image quality, yet existing methods face challenges such as high computational demands and poor generalizability to different datasets. To overcome these limitations, we propose DeepSparse, the first foundation model for sparse-view CBCT reconstruction, featuring DiCE (Dual-Dimensional Cross-Scale Embedding), a novel network that integrates multi-view 2D features and multi-scale 3D features. Additionally, we introduce the HyViP (Hybrid View Sampling Pretraining) framework, which pretrains the model on large datasets with both sparse-view and dense-view projections, and a two-step finetuning strategy to adapt and refine the model for new datasets. Extensive experiments and ablation studies demonstrate that our proposed DeepSparse achieves superior reconstruction quality compared to state-of-the-art methods, paving the way for safer and more efficient CBCT imaging.
Test-Time Domain Generalization via Universe Learning: A Multi-Graph Matching Approach for Medical Image Segmentation
Mar 17, 2025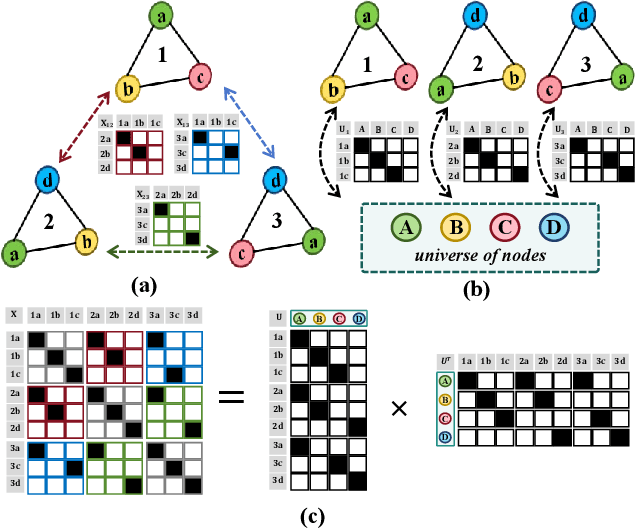
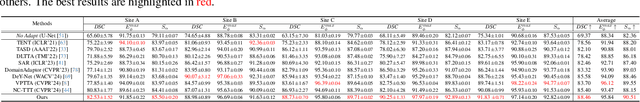
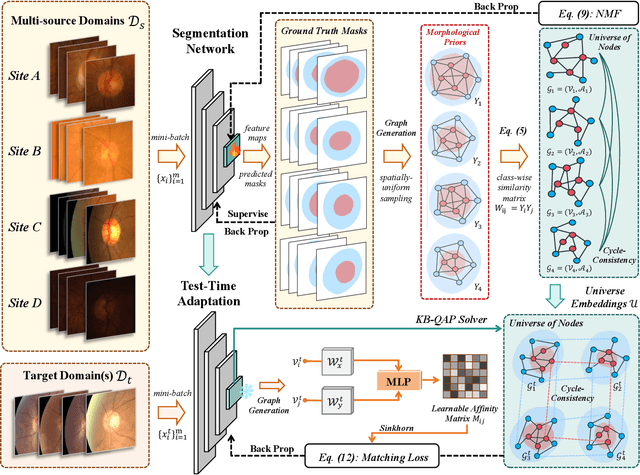
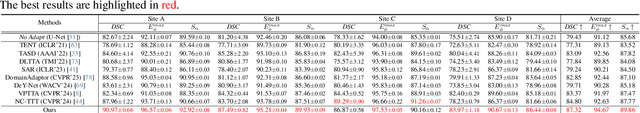
Abstract:Despite domain generalization (DG) has significantly addressed the performance degradation of pre-trained models caused by domain shifts, it often falls short in real-world deployment. Test-time adaptation (TTA), which adjusts a learned model using unlabeled test data, presents a promising solution. However, most existing TTA methods struggle to deliver strong performance in medical image segmentation, primarily because they overlook the crucial prior knowledge inherent to medical images. To address this challenge, we incorporate morphological information and propose a framework based on multi-graph matching. Specifically, we introduce learnable universe embeddings that integrate morphological priors during multi-source training, along with novel unsupervised test-time paradigms for domain adaptation. This approach guarantees cycle-consistency in multi-matching while enabling the model to more effectively capture the invariant priors of unseen data, significantly mitigating the effects of domain shifts. Extensive experiments demonstrate that our method outperforms other state-of-the-art approaches on two medical image segmentation benchmarks for both multi-source and single-source domain generalization tasks. The source code is available at https://github.com/Yore0/TTDG-MGM.
Bidirectional Recurrence for Cardiac Motion Tracking with Gaussian Process Latent Coding
Oct 28, 2024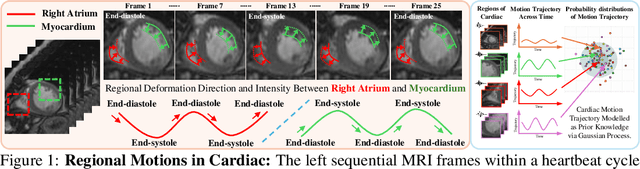
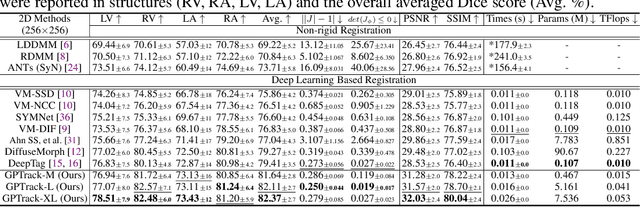
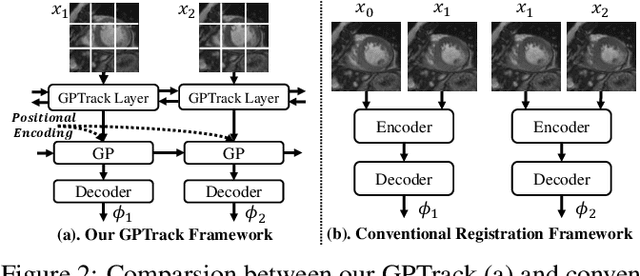
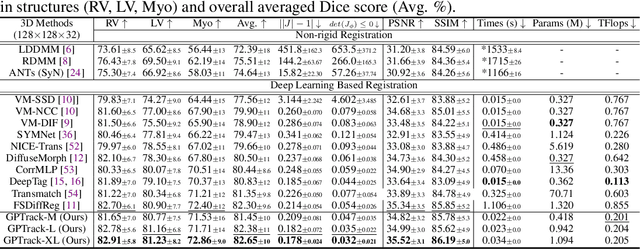
Abstract:Quantitative analysis of cardiac motion is crucial for assessing cardiac function. This analysis typically uses imaging modalities such as MRI and Echocardiograms that capture detailed image sequences throughout the heartbeat cycle. Previous methods predominantly focused on the analysis of image pairs lacking consideration of the motion dynamics and spatial variability. Consequently, these methods often overlook the long-term relationships and regional motion characteristic of cardiac. To overcome these limitations, we introduce the \textbf{GPTrack}, a novel unsupervised framework crafted to fully explore the temporal and spatial dynamics of cardiac motion. The GPTrack enhances motion tracking by employing the sequential Gaussian Process in the latent space and encoding statistics by spatial information at each time stamp, which robustly promotes temporal consistency and spatial variability of cardiac dynamics. Also, we innovatively aggregate sequential information in a bidirectional recursive manner, mimicking the behavior of diffeomorphic registration to better capture consistent long-term relationships of motions across cardiac regions such as the ventricles and atria. Our GPTrack significantly improves the precision of motion tracking in both 3D and 4D medical images while maintaining computational efficiency. The code is available at: https://github.com/xmed-lab/GPTrack
CardiacNet: Learning to Reconstruct Abnormalities for Cardiac Disease Assessment from Echocardiogram Videos
Oct 28, 2024Abstract:Echocardiogram video plays a crucial role in analysing cardiac function and diagnosing cardiac diseases. Current deep neural network methods primarily aim to enhance diagnosis accuracy by incorporating prior knowledge, such as segmenting cardiac structures or lesions annotated by human experts. However, diagnosing the inconsistent behaviours of the heart, which exist across both spatial and temporal dimensions, remains extremely challenging. For instance, the analysis of cardiac motion acquires both spatial and temporal information from the heartbeat cycle. To address this issue, we propose a novel reconstruction-based approach named CardiacNet to learn a better representation of local cardiac structures and motion abnormalities through echocardiogram videos. CardiacNet is accompanied by the Consistency Deformation Codebook (CDC) and the Consistency Deformed-Discriminator (CDD) to learn the commonalities across abnormal and normal samples by incorporating cardiac prior knowledge. In addition, we propose benchmark datasets named CardiacNet-PAH and CardiacNet-ASD to evaluate the effectiveness of cardiac disease assessment. In experiments, our CardiacNet can achieve state-of-the-art results in three different cardiac disease assessment tasks on public datasets CAMUS, EchoNet, and our datasets. The code and dataset are available at: https://github.com/xmed-lab/CardiacNet.
C^2RV: Cross-Regional and Cross-View Learning for Sparse-View CBCT Reconstruction
Jun 06, 2024Abstract:Cone beam computed tomography (CBCT) is an important imaging technology widely used in medical scenarios, such as diagnosis and preoperative planning. Using fewer projection views to reconstruct CT, also known as sparse-view reconstruction, can reduce ionizing radiation and further benefit interventional radiology. Compared with sparse-view reconstruction for traditional parallel/fan-beam CT, CBCT reconstruction is more challenging due to the increased dimensionality caused by the measurement process based on cone-shaped X-ray beams. As a 2D-to-3D reconstruction problem, although implicit neural representations have been introduced to enable efficient training, only local features are considered and different views are processed equally in previous works, resulting in spatial inconsistency and poor performance on complicated anatomies. To this end, we propose C^2RV by leveraging explicit multi-scale volumetric representations to enable cross-regional learning in the 3D space. Additionally, the scale-view cross-attention module is introduced to adaptively aggregate multi-scale and multi-view features. Extensive experiments demonstrate that our C^2RV achieves consistent and significant improvement over previous state-of-the-art methods on datasets with diverse anatomy.
DiffCMR: Fast Cardiac MRI Reconstruction with Diffusion Probabilistic Models
Dec 08, 2023



Abstract:Performing magnetic resonance imaging (MRI) reconstruction from under-sampled k-space data can accelerate the procedure to acquire MRI scans and reduce patients' discomfort. The reconstruction problem is usually formulated as a denoising task that removes the noise in under-sampled MRI image slices. Although previous GAN-based methods have achieved good performance in image denoising, they are difficult to train and require careful tuning of hyperparameters. In this paper, we propose a novel MRI denoising framework DiffCMR by leveraging conditional denoising diffusion probabilistic models. Specifically, DiffCMR perceives conditioning signals from the under-sampled MRI image slice and generates its corresponding fully-sampled MRI image slice. During inference, we adopt a multi-round ensembling strategy to stabilize the performance. We validate DiffCMR with cine reconstruction and T1/T2 mapping tasks on MICCAI 2023 Cardiac MRI Reconstruction Challenge (CMRxRecon) dataset. Results show that our method achieves state-of-the-art performance, exceeding previous methods by a significant margin. Code is available at https://github.com/xmed-lab/DiffCMR.
GraphEcho: Graph-Driven Unsupervised Domain Adaptation for Echocardiogram Video Segmentation
Sep 20, 2023



Abstract:Echocardiogram video segmentation plays an important role in cardiac disease diagnosis. This paper studies the unsupervised domain adaption (UDA) for echocardiogram video segmentation, where the goal is to generalize the model trained on the source domain to other unlabelled target domains. Existing UDA segmentation methods are not suitable for this task because they do not model local information and the cyclical consistency of heartbeat. In this paper, we introduce a newly collected CardiacUDA dataset and a novel GraphEcho method for cardiac structure segmentation. Our GraphEcho comprises two innovative modules, the Spatial-wise Cross-domain Graph Matching (SCGM) and the Temporal Cycle Consistency (TCC) module, which utilize prior knowledge of echocardiogram videos, i.e., consistent cardiac structure across patients and centers and the heartbeat cyclical consistency, respectively. These two modules can better align global and local features from source and target domains, improving UDA segmentation results. Experimental results showed that our GraphEcho outperforms existing state-of-the-art UDA segmentation methods. Our collected dataset and code will be publicly released upon acceptance. This work will lay a new and solid cornerstone for cardiac structure segmentation from echocardiogram videos. Code and dataset are available at: https://github.com/xmed-lab/GraphEcho
GL-Fusion: Global-Local Fusion Network for Multi-view Echocardiogram Video Segmentation
Sep 20, 2023



Abstract:Cardiac structure segmentation from echocardiogram videos plays a crucial role in diagnosing heart disease. The combination of multi-view echocardiogram data is essential to enhance the accuracy and robustness of automated methods. However, due to the visual disparity of the data, deriving cross-view context information remains a challenging task, and unsophisticated fusion strategies can even lower performance. In this study, we propose a novel Gobal-Local fusion (GL-Fusion) network to jointly utilize multi-view information globally and locally that improve the accuracy of echocardiogram analysis. Specifically, a Multi-view Global-based Fusion Module (MGFM) is proposed to extract global context information and to explore the cyclic relationship of different heartbeat cycles in an echocardiogram video. Additionally, a Multi-view Local-based Fusion Module (MLFM) is designed to extract correlations of cardiac structures from different views. Furthermore, we collect a multi-view echocardiogram video dataset (MvEVD) to evaluate our method. Our method achieves an 82.29% average dice score, which demonstrates a 7.83% improvement over the baseline method, and outperforms other existing state-of-the-art methods. To our knowledge, this is the first exploration of a multi-view method for echocardiogram video segmentation. Code available at: https://github.com/xmed-lab/GL-Fusion
Abandoning the Bayer-Filter to See in the Dark
Mar 22, 2022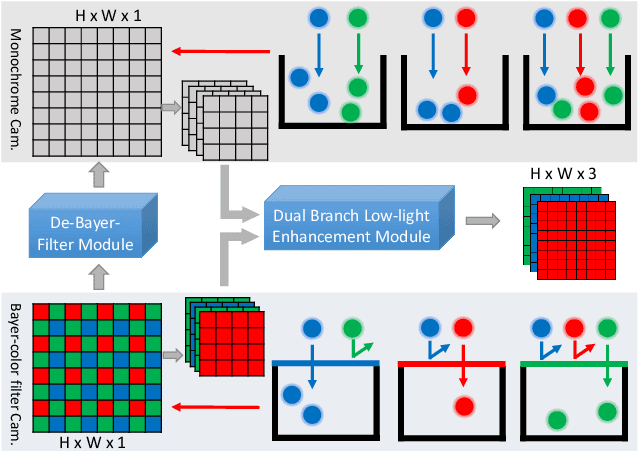
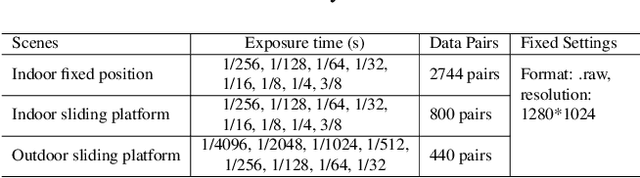
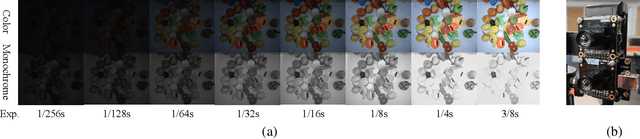
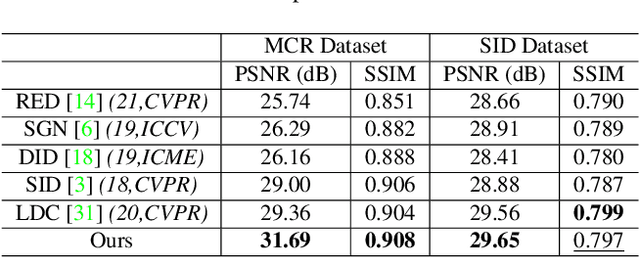
Abstract:Low-light image enhancement - a pervasive but challenging problem, plays a central role in enhancing the visibility of an image captured in a poor illumination environment. Due to the fact that not all photons can pass the Bayer-Filter on the sensor of the color camera, in this work, we first present a De-Bayer-Filter simulator based on deep neural networks to generate a monochrome raw image from the colored raw image. Next, a fully convolutional network is proposed to achieve the low-light image enhancement by fusing colored raw data with synthesized monochrome raw data. Channel-wise attention is also introduced to the fusion process to establish a complementary interaction between features from colored and monochrome raw images. To train the convolutional networks, we propose a dataset with monochrome and color raw pairs named Mono-Colored Raw paired dataset (MCR) collected by using a monochrome camera without Bayer-Filter and a color camera with Bayer-Filter. The proposed pipeline take advantages of the fusion of the virtual monochrome and the color raw images and our extensive experiments indicate that significant improvement can be achieved by leveraging raw sensor data and data-driven learning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge