Yiqun Lin
Cross-view Generalized Diffusion Model for Sparse-view CT Reconstruction
Aug 14, 2025Abstract:Sparse-view computed tomography (CT) reduces radiation exposure by subsampling projection views, but conventional reconstruction methods produce severe streak artifacts with undersampled data. While deep-learning-based methods enable single-step artifact suppression, they often produce over-smoothed results under significant sparsity. Though diffusion models improve reconstruction via iterative refinement and generative priors, they require hundreds of sampling steps and struggle with stability in highly sparse regimes. To tackle these concerns, we present the Cross-view Generalized Diffusion Model (CvG-Diff), which reformulates sparse-view CT reconstruction as a generalized diffusion process. Unlike existing diffusion approaches that rely on stochastic Gaussian degradation, CvG-Diff explicitly models image-domain artifacts caused by angular subsampling as a deterministic degradation operator, leveraging correlations across sparse-view CT at different sample rates. To address the inherent artifact propagation and inefficiency of sequential sampling in generalized diffusion model, we introduce two innovations: Error-Propagating Composite Training (EPCT), which facilitates identifying error-prone regions and suppresses propagated artifacts, and Semantic-Prioritized Dual-Phase Sampling (SPDPS), an adaptive strategy that prioritizes semantic correctness before detail refinement. Together, these innovations enable CvG-Diff to achieve high-quality reconstructions with minimal iterations, achieving 38.34 dB PSNR and 0.9518 SSIM for 18-view CT using only \textbf{10} steps on AAPM-LDCT dataset. Extensive experiments demonstrate the superiority of CvG-Diff over state-of-the-art sparse-view CT reconstruction methods. The code is available at https://github.com/xmed-lab/CvG-Diff.
DeepSparse: A Foundation Model for Sparse-View CBCT Reconstruction
May 05, 2025Abstract:Cone-beam computed tomography (CBCT) is a critical 3D imaging technology in the medical field, while the high radiation exposure required for high-quality imaging raises significant concerns, particularly for vulnerable populations. Sparse-view reconstruction reduces radiation by using fewer X-ray projections while maintaining image quality, yet existing methods face challenges such as high computational demands and poor generalizability to different datasets. To overcome these limitations, we propose DeepSparse, the first foundation model for sparse-view CBCT reconstruction, featuring DiCE (Dual-Dimensional Cross-Scale Embedding), a novel network that integrates multi-view 2D features and multi-scale 3D features. Additionally, we introduce the HyViP (Hybrid View Sampling Pretraining) framework, which pretrains the model on large datasets with both sparse-view and dense-view projections, and a two-step finetuning strategy to adapt and refine the model for new datasets. Extensive experiments and ablation studies demonstrate that our proposed DeepSparse achieves superior reconstruction quality compared to state-of-the-art methods, paving the way for safer and more efficient CBCT imaging.
Bidirectional Recurrence for Cardiac Motion Tracking with Gaussian Process Latent Coding
Oct 28, 2024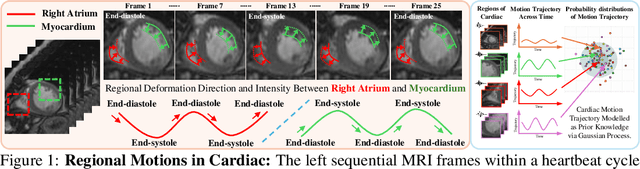
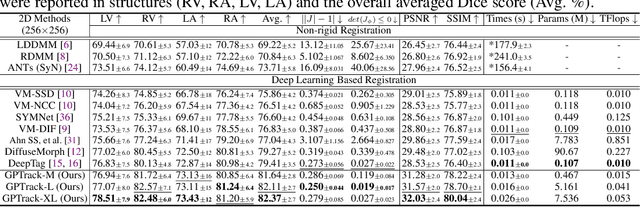
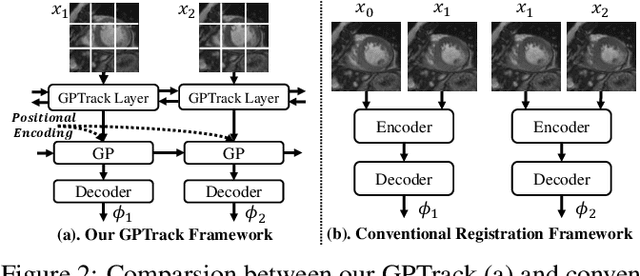
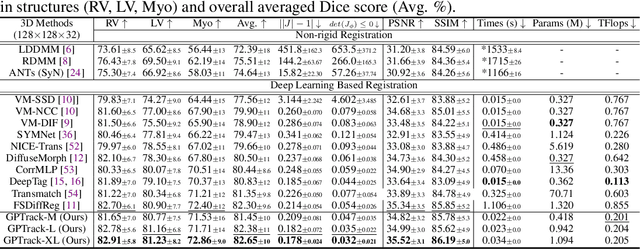
Abstract:Quantitative analysis of cardiac motion is crucial for assessing cardiac function. This analysis typically uses imaging modalities such as MRI and Echocardiograms that capture detailed image sequences throughout the heartbeat cycle. Previous methods predominantly focused on the analysis of image pairs lacking consideration of the motion dynamics and spatial variability. Consequently, these methods often overlook the long-term relationships and regional motion characteristic of cardiac. To overcome these limitations, we introduce the \textbf{GPTrack}, a novel unsupervised framework crafted to fully explore the temporal and spatial dynamics of cardiac motion. The GPTrack enhances motion tracking by employing the sequential Gaussian Process in the latent space and encoding statistics by spatial information at each time stamp, which robustly promotes temporal consistency and spatial variability of cardiac dynamics. Also, we innovatively aggregate sequential information in a bidirectional recursive manner, mimicking the behavior of diffeomorphic registration to better capture consistent long-term relationships of motions across cardiac regions such as the ventricles and atria. Our GPTrack significantly improves the precision of motion tracking in both 3D and 4D medical images while maintaining computational efficiency. The code is available at: https://github.com/xmed-lab/GPTrack
CardiacNet: Learning to Reconstruct Abnormalities for Cardiac Disease Assessment from Echocardiogram Videos
Oct 28, 2024Abstract:Echocardiogram video plays a crucial role in analysing cardiac function and diagnosing cardiac diseases. Current deep neural network methods primarily aim to enhance diagnosis accuracy by incorporating prior knowledge, such as segmenting cardiac structures or lesions annotated by human experts. However, diagnosing the inconsistent behaviours of the heart, which exist across both spatial and temporal dimensions, remains extremely challenging. For instance, the analysis of cardiac motion acquires both spatial and temporal information from the heartbeat cycle. To address this issue, we propose a novel reconstruction-based approach named CardiacNet to learn a better representation of local cardiac structures and motion abnormalities through echocardiogram videos. CardiacNet is accompanied by the Consistency Deformation Codebook (CDC) and the Consistency Deformed-Discriminator (CDD) to learn the commonalities across abnormal and normal samples by incorporating cardiac prior knowledge. In addition, we propose benchmark datasets named CardiacNet-PAH and CardiacNet-ASD to evaluate the effectiveness of cardiac disease assessment. In experiments, our CardiacNet can achieve state-of-the-art results in three different cardiac disease assessment tasks on public datasets CAMUS, EchoNet, and our datasets. The code and dataset are available at: https://github.com/xmed-lab/CardiacNet.
Tri-Plane Mamba: Efficiently Adapting Segment Anything Model for 3D Medical Images
Sep 13, 2024Abstract:General networks for 3D medical image segmentation have recently undergone extensive exploration. Behind the exceptional performance of these networks lies a significant demand for a large volume of pixel-level annotated data, which is time-consuming and labor-intensive. The emergence of the Segment Anything Model (SAM) has enabled this model to achieve superior performance in 2D medical image segmentation tasks via parameter- and data-efficient feature adaptation. However, the introduction of additional depth channels in 3D medical images not only prevents the sharing of 2D pre-trained features but also results in a quadratic increase in the computational cost for adapting SAM. To overcome these challenges, we present the Tri-Plane Mamba (TP-Mamba) adapters tailored for the SAM, featuring two major innovations: 1) multi-scale 3D convolutional adapters, optimized for efficiently processing local depth-level information, 2) a tri-plane mamba module, engineered to capture long-range depth-level representation without significantly increasing computational costs. This approach achieves state-of-the-art performance in 3D CT organ segmentation tasks. Remarkably, this superior performance is maintained even with scarce training data. Specifically using only three CT training samples from the BTCV dataset, it surpasses conventional 3D segmentation networks, attaining a Dice score that is up to 12% higher.
Spatial-Division Augmented Occupancy Field for Bone Shape Reconstruction from Biplanar X-Rays
Jul 22, 2024



Abstract:Retrieving 3D bone anatomy from biplanar X-ray images is crucial since it can significantly reduce radiation exposure compared to traditional CT-based methods. Although various deep learning models have been proposed to address this complex task, they suffer from two limitations: 1) They employ voxel representation for bone shape and exploit 3D convolutional layers to capture anatomy prior, which are memory-intensive and limit the reconstruction resolution. 2) They overlook the prevalent occlusion effect within X-ray images and directly extract features using a simple loss, which struggles to fully exploit complex X-ray information. To tackle these concerns, we present Spatial-division Augmented Occupancy Field~(SdAOF). SdAOF adopts the continuous occupancy field for shape representation, reformulating the reconstruction problem as a per-point occupancy value prediction task. Its implicit and continuous nature enables memory-efficient training and fine-scale surface reconstruction at different resolutions during the inference. Moreover, we propose a novel spatial-division augmented distillation strategy to provide feature-level guidance for capturing the occlusion relationship. Extensive experiments on the pelvis reconstruction dataset show that SdAOF outperforms state-of-the-art methods and reconstructs fine-scale bone surfaces.The code is available at https://github.com/xmed-lab/SdAOF
Learning 3D Gaussians for Extremely Sparse-View Cone-Beam CT Reconstruction
Jul 01, 2024Abstract:Cone-Beam Computed Tomography (CBCT) is an indispensable technique in medical imaging, yet the associated radiation exposure raises concerns in clinical practice. To mitigate these risks, sparse-view reconstruction has emerged as an essential research direction, aiming to reduce the radiation dose by utilizing fewer projections for CT reconstruction. Although implicit neural representations have been introduced for sparse-view CBCT reconstruction, existing methods primarily focus on local 2D features queried from sparse projections, which is insufficient to process the more complicated anatomical structures, such as the chest. To this end, we propose a novel reconstruction framework, namely DIF-Gaussian, which leverages 3D Gaussians to represent the feature distribution in the 3D space, offering additional 3D spatial information to facilitate the estimation of attenuation coefficients. Furthermore, we incorporate test-time optimization during inference to further improve the generalization capability of the model. We evaluate DIF-Gaussian on two public datasets, showing significantly superior reconstruction performance than previous state-of-the-art methods.
C^2RV: Cross-Regional and Cross-View Learning for Sparse-View CBCT Reconstruction
Jun 06, 2024Abstract:Cone beam computed tomography (CBCT) is an important imaging technology widely used in medical scenarios, such as diagnosis and preoperative planning. Using fewer projection views to reconstruct CT, also known as sparse-view reconstruction, can reduce ionizing radiation and further benefit interventional radiology. Compared with sparse-view reconstruction for traditional parallel/fan-beam CT, CBCT reconstruction is more challenging due to the increased dimensionality caused by the measurement process based on cone-shaped X-ray beams. As a 2D-to-3D reconstruction problem, although implicit neural representations have been introduced to enable efficient training, only local features are considered and different views are processed equally in previous works, resulting in spatial inconsistency and poor performance on complicated anatomies. To this end, we propose C^2RV by leveraging explicit multi-scale volumetric representations to enable cross-regional learning in the 3D space. Additionally, the scale-view cross-attention module is introduced to adaptively aggregate multi-scale and multi-view features. Extensive experiments demonstrate that our C^2RV achieves consistent and significant improvement over previous state-of-the-art methods on datasets with diverse anatomy.
Exploiting Hierarchical Interactions for Protein Surface Learning
Jan 17, 2024Abstract:Predicting interactions between proteins is one of the most important yet challenging problems in structural bioinformatics. Intrinsically, potential function sites in protein surfaces are determined by both geometric and chemical features. However, existing works only consider handcrafted or individually learned chemical features from the atom type and extract geometric features independently. Here, we identify two key properties of effective protein surface learning: 1) relationship among atoms: atoms are linked with each other by covalent bonds to form biomolecules instead of appearing alone, leading to the significance of modeling the relationship among atoms in chemical feature learning. 2) hierarchical feature interaction: the neighboring residue effect validates the significance of hierarchical feature interaction among atoms and between surface points and atoms (or residues). In this paper, we present a principled framework based on deep learning techniques, namely Hierarchical Chemical and Geometric Feature Interaction Network (HCGNet), for protein surface analysis by bridging chemical and geometric features with hierarchical interactions. Extensive experiments demonstrate that our method outperforms the prior state-of-the-art method by 2.3% in site prediction task and 3.2% in interaction matching task, respectively. Our code is available at https://github.com/xmed-lab/HCGNet.
DiffCMR: Fast Cardiac MRI Reconstruction with Diffusion Probabilistic Models
Dec 08, 2023



Abstract:Performing magnetic resonance imaging (MRI) reconstruction from under-sampled k-space data can accelerate the procedure to acquire MRI scans and reduce patients' discomfort. The reconstruction problem is usually formulated as a denoising task that removes the noise in under-sampled MRI image slices. Although previous GAN-based methods have achieved good performance in image denoising, they are difficult to train and require careful tuning of hyperparameters. In this paper, we propose a novel MRI denoising framework DiffCMR by leveraging conditional denoising diffusion probabilistic models. Specifically, DiffCMR perceives conditioning signals from the under-sampled MRI image slice and generates its corresponding fully-sampled MRI image slice. During inference, we adopt a multi-round ensembling strategy to stabilize the performance. We validate DiffCMR with cine reconstruction and T1/T2 mapping tasks on MICCAI 2023 Cardiac MRI Reconstruction Challenge (CMRxRecon) dataset. Results show that our method achieves state-of-the-art performance, exceeding previous methods by a significant margin. Code is available at https://github.com/xmed-lab/DiffCMR.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge