Jiarong Guo
AI-Enabled Accurate Non-Invasive Assessment of Pulmonary Hypertension Progression via Multi-Modal Echocardiography
May 12, 2025Abstract:Echocardiographers can detect pulmonary hypertension using Doppler echocardiography; however, accurately assessing its progression often proves challenging. Right heart catheterization (RHC), the gold standard for precise evaluation, is invasive and unsuitable for routine use, limiting its practicality for timely diagnosis and monitoring of pulmonary hypertension progression. Here, we propose MePH, a multi-view, multi-modal vision-language model to accurately assess pulmonary hypertension progression using non-invasive echocardiography. We constructed a large dataset comprising paired standardized echocardiogram videos, spectral images and RHC data, covering 1,237 patient cases from 12 medical centers. For the first time, MePH precisely models the correlation between non-invasive multi-view, multi-modal echocardiography and the pressure and resistance obtained via RHC. We show that MePH significantly outperforms echocardiographers' assessments using echocardiography, reducing the mean absolute error in estimating mean pulmonary arterial pressure (mPAP) and pulmonary vascular resistance (PVR) by 49.73% and 43.81%, respectively. In eight independent external hospitals, MePH achieved a mean absolute error of 3.147 for PVR assessment. Furthermore, MePH achieved an area under the curve of 0.921, surpassing echocardiographers (area under the curve of 0.842) in accurately predicting the severity of pulmonary hypertension, whether mild or severe. A prospective study demonstrated that MePH can predict treatment efficacy for patients. Our work provides pulmonary hypertension patients with a non-invasive and timely method for monitoring disease progression, improving the accuracy and efficiency of pulmonary hypertension management while enabling earlier interventions and more personalized treatment decisions.
DeepSparse: A Foundation Model for Sparse-View CBCT Reconstruction
May 05, 2025Abstract:Cone-beam computed tomography (CBCT) is a critical 3D imaging technology in the medical field, while the high radiation exposure required for high-quality imaging raises significant concerns, particularly for vulnerable populations. Sparse-view reconstruction reduces radiation by using fewer X-ray projections while maintaining image quality, yet existing methods face challenges such as high computational demands and poor generalizability to different datasets. To overcome these limitations, we propose DeepSparse, the first foundation model for sparse-view CBCT reconstruction, featuring DiCE (Dual-Dimensional Cross-Scale Embedding), a novel network that integrates multi-view 2D features and multi-scale 3D features. Additionally, we introduce the HyViP (Hybrid View Sampling Pretraining) framework, which pretrains the model on large datasets with both sparse-view and dense-view projections, and a two-step finetuning strategy to adapt and refine the model for new datasets. Extensive experiments and ablation studies demonstrate that our proposed DeepSparse achieves superior reconstruction quality compared to state-of-the-art methods, paving the way for safer and more efficient CBCT imaging.
CardiacNet: Learning to Reconstruct Abnormalities for Cardiac Disease Assessment from Echocardiogram Videos
Oct 28, 2024Abstract:Echocardiogram video plays a crucial role in analysing cardiac function and diagnosing cardiac diseases. Current deep neural network methods primarily aim to enhance diagnosis accuracy by incorporating prior knowledge, such as segmenting cardiac structures or lesions annotated by human experts. However, diagnosing the inconsistent behaviours of the heart, which exist across both spatial and temporal dimensions, remains extremely challenging. For instance, the analysis of cardiac motion acquires both spatial and temporal information from the heartbeat cycle. To address this issue, we propose a novel reconstruction-based approach named CardiacNet to learn a better representation of local cardiac structures and motion abnormalities through echocardiogram videos. CardiacNet is accompanied by the Consistency Deformation Codebook (CDC) and the Consistency Deformed-Discriminator (CDD) to learn the commonalities across abnormal and normal samples by incorporating cardiac prior knowledge. In addition, we propose benchmark datasets named CardiacNet-PAH and CardiacNet-ASD to evaluate the effectiveness of cardiac disease assessment. In experiments, our CardiacNet can achieve state-of-the-art results in three different cardiac disease assessment tasks on public datasets CAMUS, EchoNet, and our datasets. The code and dataset are available at: https://github.com/xmed-lab/CardiacNet.
Self-supervised Learning for Enhancing Geometrical Modeling in 3D-Aware Generative Adversarial Network
Dec 19, 2023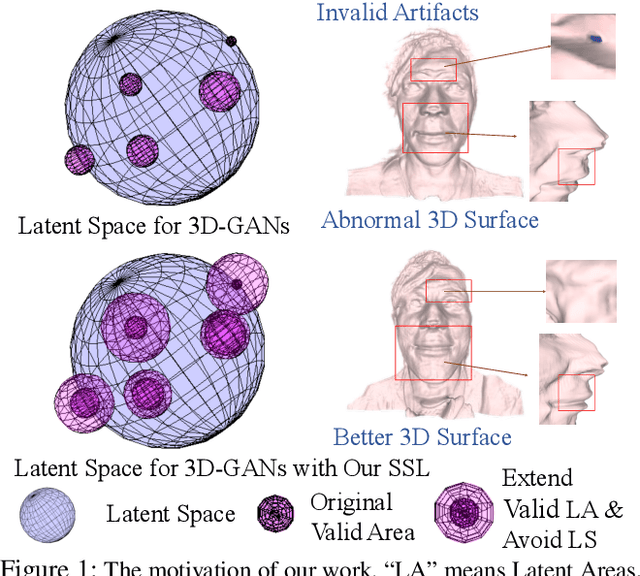
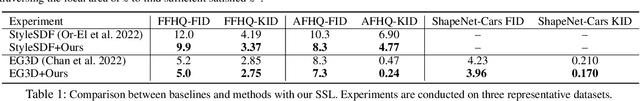
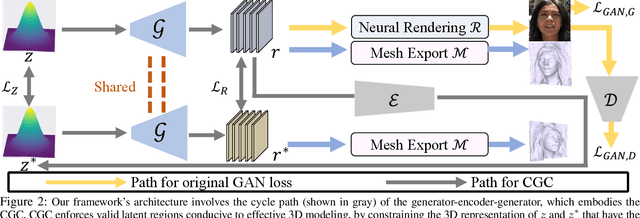
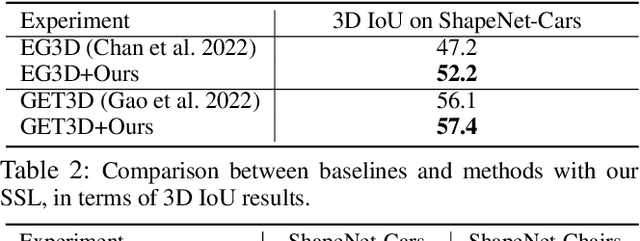
Abstract:3D-aware Generative Adversarial Networks (3D-GANs) currently exhibit artifacts in their 3D geometrical modeling, such as mesh imperfections and holes. These shortcomings are primarily attributed to the limited availability of annotated 3D data, leading to a constrained "valid latent area" for satisfactory modeling. To address this, we present a Self-Supervised Learning (SSL) technique tailored as an auxiliary loss for any 3D-GAN, designed to improve its 3D geometrical modeling capabilities. Our approach pioneers an inversion technique for 3D-GANs, integrating an encoder that performs adaptive spatially-varying range operations. Utilizing this inversion, we introduce the Cyclic Generative Constraint (CGC), aiming to densify the valid latent space. The CGC operates via augmented local latent vectors that maintain the same geometric form, and it imposes constraints on the cycle path outputs, specifically the generator-encoder-generator sequence. This SSL methodology seamlessly integrates with the inherent GAN loss, ensuring the integrity of pre-existing 3D-GAN architectures without necessitating alterations. We validate our approach with comprehensive experiments across various datasets and architectures, underscoring its efficacy. Our project website: https://3dgan-ssl.github.io
Semi-Supervised Domain Generalization for Cardiac Magnetic Resonance Image Segmentation with High Quality Pseudo Labels
Sep 30, 2022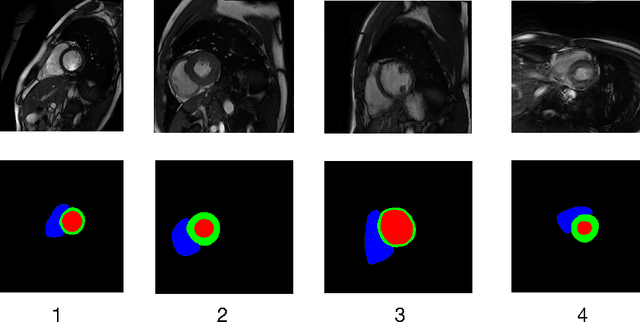
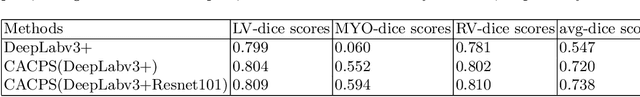
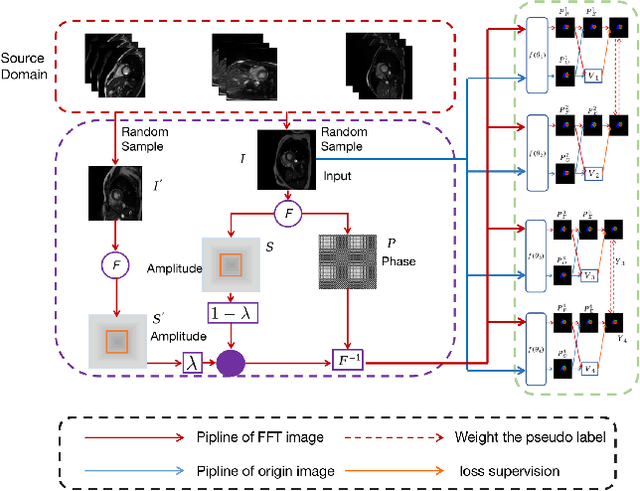
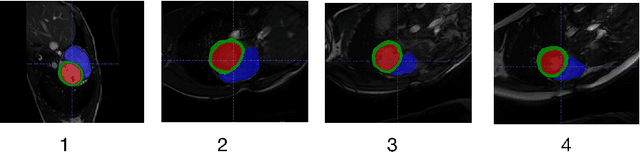
Abstract:Developing a deep learning method for medical segmentation tasks heavily relies on a large amount of labeled data. However, the annotations require professional knowledge and are limited in number. Recently, semi-supervised learning has demonstrated great potential in medical segmentation tasks. Most existing methods related to cardiac magnetic resonance images only focus on regular images with similar domains and high image quality. A semi-supervised domain generalization method was developed in [2], which enhances the quality of pseudo labels on varied datasets. In this paper, we follow the strategy in [2] and present a domain generalization method for semi-supervised medical segmentation. Our main goal is to improve the quality of pseudo labels under extreme MRI Analysis with various domains. We perform Fourier transformation on input images to learn low-level statistics and cross-domain information. Then we feed the augmented images as input to the double cross pseudo supervision networks to calculate the variance among pseudo labels. We evaluate our method on the CMRxMotion dataset [1]. With only partially labeled data and without domain labels, our approach consistently generates accurate segmentation results of cardiac magnetic resonance images with different respiratory motions. Code will be available after the conference.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge