Xiaowei Xu
FlowRoI A Fast Optical Flow Driven Region of Interest Extraction Framework for High-Throughput Image Compression in Immune Cell Migration Analysis
Nov 18, 2025Abstract:Autonomous migration is essential for the function of immune cells such as neutrophils and plays a pivotal role in diverse diseases. Recently, we introduced ComplexEye, a multi-lens array microscope comprising 16 independent aberration-corrected glass lenses arranged at the pitch of a 96-well plate, capable of capturing high-resolution movies of migrating cells. This architecture enables high-throughput live-cell video microscopy for migration analysis, supporting routine quantification of autonomous motility with strong potential for clinical translation. However, ComplexEye and similar high-throughput imaging platforms generate data at an exponential rate, imposing substantial burdens on storage and transmission. To address this challenge, we present FlowRoI, a fast optical-flow-based region of interest (RoI) extraction framework designed for high-throughput image compression in immune cell migration studies. FlowRoI estimates optical flow between consecutive frames and derives RoI masks that reliably cover nearly all migrating cells. The raw image and its corresponding RoI mask are then jointly encoded using JPEG2000 to enable RoI-aware compression. FlowRoI operates with high computational efficiency, achieving runtimes comparable to standard JPEG2000 and reaching an average throughput of about 30 frames per second on a modern laptop equipped with an Intel i7-1255U CPU. In terms of image quality, FlowRoI yields higher peak signal-to-noise ratio (PSNR) in cellular regions and achieves 2.0-2.2x higher compression rates at matched PSNR compared to standard JPEG2000.
PathMR: Multimodal Visual Reasoning for Interpretable Pathology Diagnosis
Aug 28, 2025Abstract:Deep learning based automated pathological diagnosis has markedly improved diagnostic efficiency and reduced variability between observers, yet its clinical adoption remains limited by opaque model decisions and a lack of traceable rationale. To address this, recent multimodal visual reasoning architectures provide a unified framework that generates segmentation masks at the pixel level alongside semantically aligned textual explanations. By localizing lesion regions and producing expert style diagnostic narratives, these models deliver the transparent and interpretable insights necessary for dependable AI assisted pathology. Building on these advancements, we propose PathMR, a cell-level Multimodal visual Reasoning framework for Pathological image analysis. Given a pathological image and a textual query, PathMR generates expert-level diagnostic explanations while simultaneously predicting cell distribution patterns. To benchmark its performance, we evaluated our approach on the publicly available PathGen dataset as well as on our newly developed GADVR dataset. Extensive experiments on these two datasets demonstrate that PathMR consistently outperforms state-of-the-art visual reasoning methods in text generation quality, segmentation accuracy, and cross-modal alignment. These results highlight the potential of PathMR for improving interpretability in AI-driven pathological diagnosis. The code will be publicly available in https://github.com/zhangye-zoe/PathMR.
AI-Enabled Accurate Non-Invasive Assessment of Pulmonary Hypertension Progression via Multi-Modal Echocardiography
May 12, 2025Abstract:Echocardiographers can detect pulmonary hypertension using Doppler echocardiography; however, accurately assessing its progression often proves challenging. Right heart catheterization (RHC), the gold standard for precise evaluation, is invasive and unsuitable for routine use, limiting its practicality for timely diagnosis and monitoring of pulmonary hypertension progression. Here, we propose MePH, a multi-view, multi-modal vision-language model to accurately assess pulmonary hypertension progression using non-invasive echocardiography. We constructed a large dataset comprising paired standardized echocardiogram videos, spectral images and RHC data, covering 1,237 patient cases from 12 medical centers. For the first time, MePH precisely models the correlation between non-invasive multi-view, multi-modal echocardiography and the pressure and resistance obtained via RHC. We show that MePH significantly outperforms echocardiographers' assessments using echocardiography, reducing the mean absolute error in estimating mean pulmonary arterial pressure (mPAP) and pulmonary vascular resistance (PVR) by 49.73% and 43.81%, respectively. In eight independent external hospitals, MePH achieved a mean absolute error of 3.147 for PVR assessment. Furthermore, MePH achieved an area under the curve of 0.921, surpassing echocardiographers (area under the curve of 0.842) in accurately predicting the severity of pulmonary hypertension, whether mild or severe. A prospective study demonstrated that MePH can predict treatment efficacy for patients. Our work provides pulmonary hypertension patients with a non-invasive and timely method for monitoring disease progression, improving the accuracy and efficiency of pulmonary hypertension management while enabling earlier interventions and more personalized treatment decisions.
ScaleMAI: Accelerating the Development of Trusted Datasets and AI Models
Jan 06, 2025

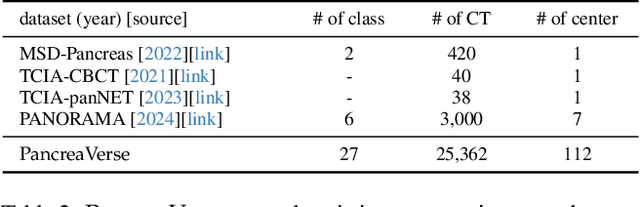

Abstract:Building trusted datasets is critical for transparent and responsible Medical AI (MAI) research, but creating even small, high-quality datasets can take years of effort from multidisciplinary teams. This process often delays AI benefits, as human-centric data creation and AI-centric model development are treated as separate, sequential steps. To overcome this, we propose ScaleMAI, an agent of AI-integrated data curation and annotation, allowing data quality and AI performance to improve in a self-reinforcing cycle and reducing development time from years to months. We adopt pancreatic tumor detection as an example. First, ScaleMAI progressively creates a dataset of 25,362 CT scans, including per-voxel annotations for benign/malignant tumors and 24 anatomical structures. Second, through progressive human-in-the-loop iterations, ScaleMAI provides Flagship AI Model that can approach the proficiency of expert annotators (30-year experience) in detecting pancreatic tumors. Flagship Model significantly outperforms models developed from smaller, fixed-quality datasets, with substantial gains in tumor detection (+14%), segmentation (+5%), and classification (72%) on three prestigious benchmarks. In summary, ScaleMAI transforms the speed, scale, and reliability of medical dataset creation, paving the way for a variety of impactful, data-driven applications.
MPBD-LSTM: A Predictive Model for Colorectal Liver Metastases Using Time Series Multi-phase Contrast-Enhanced CT Scans
Dec 02, 2024Abstract:Colorectal cancer is a prevalent form of cancer, and many patients develop colorectal cancer liver metastasis (CRLM) as a result. Early detection of CRLM is critical for improving survival rates. Radiologists usually rely on a series of multi-phase contrast-enhanced computed tomography (CECT) scans done during follow-up visits to perform early detection of the potential CRLM. These scans form unique five-dimensional data (time, phase, and axial, sagittal, and coronal planes in 3D CT). Most of the existing deep learning models can readily handle four-dimensional data (e.g., time-series 3D CT images) and it is not clear how well they can be extended to handle the additional dimension of phase. In this paper, we build a dataset of time-series CECT scans to aid in the early diagnosis of CRLM, and build upon state-of-the-art deep learning techniques to evaluate how to best predict CRLM. Our experimental results show that a multi-plane architecture based on 3D bi-directional LSTM, which we call MPBD-LSTM, works best, achieving an area under curve (AUC) of 0.79. On the other hand, analysis of the results shows that there is still great room for further improvement.
CardiacNet: Learning to Reconstruct Abnormalities for Cardiac Disease Assessment from Echocardiogram Videos
Oct 28, 2024Abstract:Echocardiogram video plays a crucial role in analysing cardiac function and diagnosing cardiac diseases. Current deep neural network methods primarily aim to enhance diagnosis accuracy by incorporating prior knowledge, such as segmenting cardiac structures or lesions annotated by human experts. However, diagnosing the inconsistent behaviours of the heart, which exist across both spatial and temporal dimensions, remains extremely challenging. For instance, the analysis of cardiac motion acquires both spatial and temporal information from the heartbeat cycle. To address this issue, we propose a novel reconstruction-based approach named CardiacNet to learn a better representation of local cardiac structures and motion abnormalities through echocardiogram videos. CardiacNet is accompanied by the Consistency Deformation Codebook (CDC) and the Consistency Deformed-Discriminator (CDD) to learn the commonalities across abnormal and normal samples by incorporating cardiac prior knowledge. In addition, we propose benchmark datasets named CardiacNet-PAH and CardiacNet-ASD to evaluate the effectiveness of cardiac disease assessment. In experiments, our CardiacNet can achieve state-of-the-art results in three different cardiac disease assessment tasks on public datasets CAMUS, EchoNet, and our datasets. The code and dataset are available at: https://github.com/xmed-lab/CardiacNet.
Contrastive Learning with Synthetic Positives
Aug 30, 2024Abstract:Contrastive learning with the nearest neighbor has proved to be one of the most efficient self-supervised learning (SSL) techniques by utilizing the similarity of multiple instances within the same class. However, its efficacy is constrained as the nearest neighbor algorithm primarily identifies ``easy'' positive pairs, where the representations are already closely located in the embedding space. In this paper, we introduce a novel approach called Contrastive Learning with Synthetic Positives (CLSP) that utilizes synthetic images, generated by an unconditional diffusion model, as the additional positives to help the model learn from diverse positives. Through feature interpolation in the diffusion model sampling process, we generate images with distinct backgrounds yet similar semantic content to the anchor image. These images are considered ``hard'' positives for the anchor image, and when included as supplementary positives in the contrastive loss, they contribute to a performance improvement of over 2\% and 1\% in linear evaluation compared to the previous NNCLR and All4One methods across multiple benchmark datasets such as CIFAR10, achieving state-of-the-art methods. On transfer learning benchmarks, CLSP outperforms existing SSL frameworks on 6 out of 8 downstream datasets. We believe CLSP establishes a valuable baseline for future SSL studies incorporating synthetic data in the training process.
Data-Algorithm-Architecture Co-Optimization for Fair Neural Networks on Skin Lesion Dataset
Jul 18, 2024Abstract:As Artificial Intelligence (AI) increasingly integrates into our daily lives, fairness has emerged as a critical concern, particularly in medical AI, where datasets often reflect inherent biases due to social factors like the underrepresentation of marginalized communities and socioeconomic barriers to data collection. Traditional approaches to mitigating these biases have focused on data augmentation and the development of fairness-aware training algorithms. However, this paper argues that the architecture of neural networks, a core component of Machine Learning (ML), plays a crucial role in ensuring fairness. We demonstrate that addressing fairness effectively requires a holistic approach that simultaneously considers data, algorithms, and architecture. Utilizing Automated ML (AutoML) technology, specifically Neural Architecture Search (NAS), we introduce a novel framework, BiaslessNAS, designed to achieve fair outcomes in analyzing skin lesion datasets. BiaslessNAS incorporates fairness considerations at every stage of the NAS process, leading to the identification of neural networks that are not only more accurate but also significantly fairer. Our experiments show that BiaslessNAS achieves a 2.55% increase in accuracy and a 65.50% improvement in fairness compared to traditional NAS methods, underscoring the importance of integrating fairness into neural network architecture for better outcomes in medical AI applications.
Invisible Backdoor Attack Through Singular Value Decomposition
Mar 18, 2024



Abstract:With the widespread application of deep learning across various domains, concerns about its security have grown significantly. Among these, backdoor attacks pose a serious security threat to deep neural networks (DNNs). In recent years, backdoor attacks on neural networks have become increasingly sophisticated, aiming to compromise the security and trustworthiness of models by implanting hidden, unauthorized functionalities or triggers, leading to misleading predictions or behaviors. To make triggers less perceptible and imperceptible, various invisible backdoor attacks have been proposed. However, most of them only consider invisibility in the spatial domain, making it easy for recent defense methods to detect the generated toxic images.To address these challenges, this paper proposes an invisible backdoor attack called DEBA. DEBA leverages the mathematical properties of Singular Value Decomposition (SVD) to embed imperceptible backdoors into models during the training phase, thereby causing them to exhibit predefined malicious behavior under specific trigger conditions. Specifically, we first perform SVD on images, and then replace the minor features of trigger images with those of clean images, using them as triggers to ensure the effectiveness of the attack. As minor features are scattered throughout the entire image, the major features of clean images are preserved, making poisoned images visually indistinguishable from clean ones. Extensive experimental evaluations demonstrate that DEBA is highly effective, maintaining high perceptual quality and a high attack success rate for poisoned images. Furthermore, we assess the performance of DEBA under existing defense measures, showing that it is robust and capable of significantly evading and resisting the effects of these defense measures.
Towards Trustable Language Models: Investigating Information Quality of Large Language Models
Jan 23, 2024Abstract:Large language models (LLM) are generating information at a rapid pace, requiring users to increasingly rely and trust the data. Despite remarkable advances of LLM, Information generated by LLM is not completely trustworthy, due to challenges in information quality. Specifically, integrity of Information quality decreases due to unreliable, biased, tokenization during pre-training of LLM. Moreover, due to decreased information quality issues, has led towards hallucination, fabricated information. Unreliable information can lead towards flawed decisions in businesses, which impacts economic activity. In this work, we introduce novel mathematical information quality evaluation of LLM, we furthermore analyze and highlight information quality challenges, scaling laws to systematically scale language models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge