Wenxuan Li
Early and Prediagnostic Detection of Pancreatic Cancer from Computed Tomography
Jan 29, 2026Abstract:Pancreatic ductal adenocarcinoma (PDAC), one of the deadliest solid malignancies, is often detected at a late and inoperable stage. Retrospective reviews of prediagnostic CT scans, when conducted by expert radiologists aware that the patient later developed PDAC, frequently reveal lesions that were previously overlooked. To help detecting these lesions earlier, we developed an automated system named ePAI (early Pancreatic cancer detection with Artificial Intelligence). It was trained on data from 1,598 patients from a single medical center. In the internal test involving 1,009 patients, ePAI achieved an area under the receiver operating characteristic curve (AUC) of 0.939-0.999, a sensitivity of 95.3%, and a specificity of 98.7% for detecting small PDAC less than 2 cm in diameter, precisely localizing PDAC as small as 2 mm. In an external test involving 7,158 patients across 6 centers, ePAI achieved an AUC of 0.918-0.945, a sensitivity of 91.5%, and a specificity of 88.0%, precisely localizing PDAC as small as 5 mm. Importantly, ePAI detected PDACs on prediagnostic CT scans obtained 3 to 36 months before clinical diagnosis that had originally been overlooked by radiologists. It successfully detected and localized PDACs in 75 of 159 patients, with a median lead time of 347 days before clinical diagnosis. Our multi-reader study showed that ePAI significantly outperformed 30 board-certified radiologists by 50.3% (P < 0.05) in sensitivity while maintaining a comparable specificity of 95.4% in detecting PDACs early and prediagnostic. These findings suggest its potential of ePAI as an assistive tool to improve early detection of pancreatic cancer.
Large-Scale Label Quality Assessment for Medical Segmentation via a Vision-Language Judge and Synthetic Data
Jan 20, 2026Abstract:Large-scale medical segmentation datasets often combine manual and pseudo-labels of uneven quality, which can compromise training and evaluation. Low-quality labels may hamper performance and make the model training less robust. To address this issue, we propose SegAE (Segmentation Assessment Engine), a lightweight vision-language model (VLM) that automatically predicts label quality across 142 anatomical structures. Trained on over four million image-label pairs with quality scores, SegAE achieves a high correlation coefficient of 0.902 with ground-truth Dice similarity and evaluates a 3D mask in 0.06s. SegAE shows several practical benefits: (I) Our analysis reveals widespread low-quality labeling across public datasets; (II) SegAE improves data efficiency and training performance in active and semi-supervised learning, reducing dataset annotation cost by one-third and quality-checking time by 70% per label. This tool provides a simple and effective solution for quality control in large-scale medical segmentation datasets. The dataset, model weights, and codes are released at https://github.com/Schuture/SegAE.
Auditing Significance, Metric Choice, and Demographic Fairness in Medical AI Challenges
Dec 22, 2025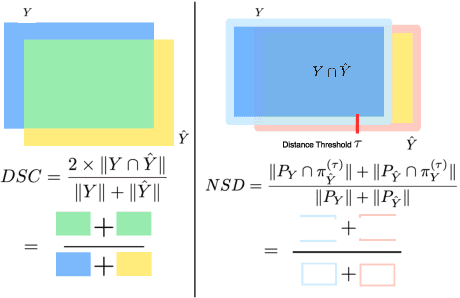
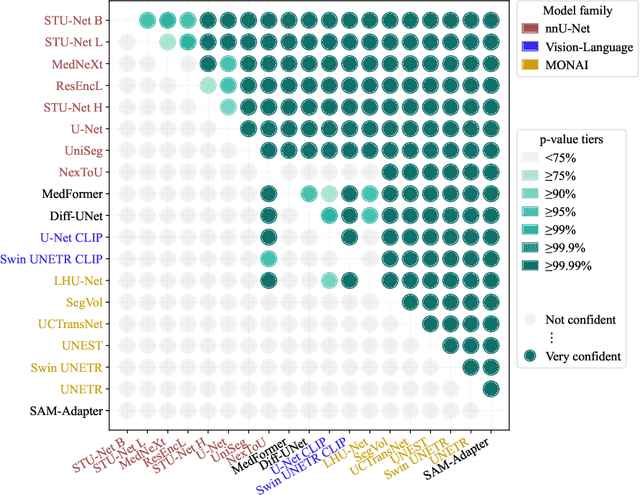
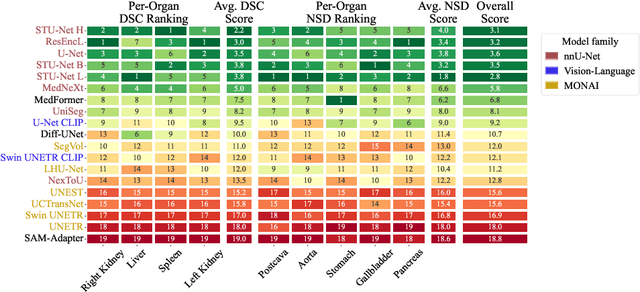
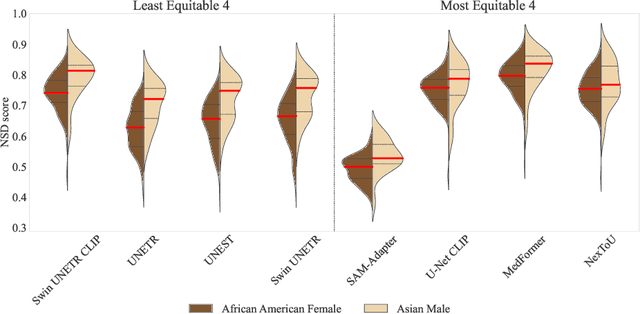
Abstract:Open challenges have become the de facto standard for comparative ranking of medical AI methods. Despite their importance, medical AI leaderboards exhibit three persistent limitations: (1) score gaps are rarely tested for statistical significance, so rank stability is unknown; (2) single averaged metrics are applied to every organ, hiding clinically important boundary errors; (3) performance across intersecting demographics is seldom reported, masking fairness and equity gaps. We introduce RankInsight, an open-source toolkit that seeks to address these limitations. RankInsight (1) computes pair-wise significance maps that show the nnU-Net family outperforms Vision-Language and MONAI submissions with high statistical certainty; (2) recomputes leaderboards with organ-appropriate metrics, reversing the order of the top four models when Dice is replaced by NSD for tubular structures; and (3) audits intersectional fairness, revealing that more than half of the MONAI-based entries have the largest gender-race discrepancy on our proprietary Johns Hopkins Hospital dataset. The RankInsight toolkit is publicly released and can be directly applied to past, ongoing, and future challenges. It enables organizers and participants to publish rankings that are statistically sound, clinically meaningful, and demographically fair.
See More, Change Less: Anatomy-Aware Diffusion for Contrast Enhancement
Dec 08, 2025Abstract:Image enhancement improves visual quality and helps reveal details that are hard to see in the original image. In medical imaging, it can support clinical decision-making, but current models often over-edit. This can distort organs, create false findings, and miss small tumors because these models do not understand anatomy or contrast dynamics. We propose SMILE, an anatomy-aware diffusion model that learns how organs are shaped and how they take up contrast. It enhances only clinically relevant regions while leaving all other areas unchanged. SMILE introduces three key ideas: (1) structure-aware supervision that follows true organ boundaries and contrast patterns; (2) registration-free learning that works directly with unaligned multi-phase CT scans; (3) unified inference that provides fast and consistent enhancement across all contrast phases. Across six external datasets, SMILE outperforms existing methods in image quality (14.2% higher SSIM, 20.6% higher PSNR, 50% better FID) and in clinical usefulness by producing anatomically accurate and diagnostically meaningful images. SMILE also improves cancer detection from non-contrast CT, raising the F1 score by up to 10 percent.
3D-ANC: Adaptive Neural Collapse for Robust 3D Point Cloud Recognition
Nov 10, 2025Abstract:Deep neural networks have recently achieved notable progress in 3D point cloud recognition, yet their vulnerability to adversarial perturbations poses critical security challenges in practical deployments. Conventional defense mechanisms struggle to address the evolving landscape of multifaceted attack patterns. Through systematic analysis of existing defenses, we identify that their unsatisfactory performance primarily originates from an entangled feature space, where adversarial attacks can be performed easily. To this end, we present 3D-ANC, a novel approach that capitalizes on the Neural Collapse (NC) mechanism to orchestrate discriminative feature learning. In particular, NC depicts where last-layer features and classifier weights jointly evolve into a simplex equiangular tight frame (ETF) arrangement, establishing maximally separable class prototypes. However, leveraging this advantage in 3D recognition confronts two substantial challenges: (1) prevalent class imbalance in point cloud datasets, and (2) complex geometric similarities between object categories. To tackle these obstacles, our solution combines an ETF-aligned classification module with an adaptive training framework consisting of representation-balanced learning (RBL) and dynamic feature direction loss (FDL). 3D-ANC seamlessly empowers existing models to develop disentangled feature spaces despite the complexity in 3D data distribution. Comprehensive evaluations state that 3D-ANC significantly improves the robustness of models with various structures on two datasets. For instance, DGCNN's classification accuracy is elevated from 27.2% to 80.9% on ModelNet40 -- a 53.7% absolute gain that surpasses leading baselines by 34.0%.
MTraining: Distributed Dynamic Sparse Attention for Efficient Ultra-Long Context Training
Oct 21, 2025Abstract:The adoption of long context windows has become a standard feature in Large Language Models (LLMs), as extended contexts significantly enhance their capacity for complex reasoning and broaden their applicability across diverse scenarios. Dynamic sparse attention is a promising approach for reducing the computational cost of long-context. However, efficiently training LLMs with dynamic sparse attention on ultra-long contexts-especially in distributed settings-remains a significant challenge, due in large part to worker- and step-level imbalance. This paper introduces MTraining, a novel distributed methodology leveraging dynamic sparse attention to enable efficient training for LLMs with ultra-long contexts. Specifically, MTraining integrates three key components: a dynamic sparse training pattern, balanced sparse ring attention, and hierarchical sparse ring attention. These components are designed to synergistically address the computational imbalance and communication overheads inherent in dynamic sparse attention mechanisms during the training of models with extensive context lengths. We demonstrate the efficacy of MTraining by training Qwen2.5-3B, successfully expanding its context window from 32K to 512K tokens on a cluster of 32 A100 GPUs. Our evaluations on a comprehensive suite of downstream tasks, including RULER, PG-19, InfiniteBench, and Needle In A Haystack, reveal that MTraining achieves up to a 6x higher training throughput while preserving model accuracy. Our code is available at https://github.com/microsoft/MInference/tree/main/MTraining.
PanTS: The Pancreatic Tumor Segmentation Dataset
Jul 02, 2025Abstract:PanTS is a large-scale, multi-institutional dataset curated to advance research in pancreatic CT analysis. It contains 36,390 CT scans from 145 medical centers, with expert-validated, voxel-wise annotations of over 993,000 anatomical structures, covering pancreatic tumors, pancreas head, body, and tail, and 24 surrounding anatomical structures such as vascular/skeletal structures and abdominal/thoracic organs. Each scan includes metadata such as patient age, sex, diagnosis, contrast phase, in-plane spacing, slice thickness, etc. AI models trained on PanTS achieve significantly better performance in pancreatic tumor detection, localization, and segmentation compared to those trained on existing public datasets. Our analysis indicates that these gains are directly attributable to the 16x larger-scale tumor annotations and indirectly supported by the 24 additional surrounding anatomical structures. As the largest and most comprehensive resource of its kind, PanTS offers a new benchmark for developing and evaluating AI models in pancreatic CT analysis.
PIN-WM: Learning Physics-INformed World Models for Non-Prehensile Manipulation
Apr 23, 2025Abstract:While non-prehensile manipulation (e.g., controlled pushing/poking) constitutes a foundational robotic skill, its learning remains challenging due to the high sensitivity to complex physical interactions involving friction and restitution. To achieve robust policy learning and generalization, we opt to learn a world model of the 3D rigid body dynamics involved in non-prehensile manipulations and use it for model-based reinforcement learning. We propose PIN-WM, a Physics-INformed World Model that enables efficient end-to-end identification of a 3D rigid body dynamical system from visual observations. Adopting differentiable physics simulation, PIN-WM can be learned with only few-shot and task-agnostic physical interaction trajectories. Further, PIN-WM is learned with observational loss induced by Gaussian Splatting without needing state estimation. To bridge Sim2Real gaps, we turn the learned PIN-WM into a group of Digital Cousins via physics-aware randomizations which perturb physics and rendering parameters to generate diverse and meaningful variations of the PIN-WM. Extensive evaluations on both simulation and real-world tests demonstrate that PIN-WM, enhanced with physics-aware digital cousins, facilitates learning robust non-prehensile manipulation skills with Sim2Real transfer, surpassing the Real2Sim2Real state-of-the-arts.
Revisiting Backdoor Attacks on Time Series Classification in the Frequency Domain
Mar 12, 2025Abstract:Time series classification (TSC) is a cornerstone of modern web applications, powering tasks such as financial data analysis, network traffic monitoring, and user behavior analysis. In recent years, deep neural networks (DNNs) have greatly enhanced the performance of TSC models in these critical domains. However, DNNs are vulnerable to backdoor attacks, where attackers can covertly implant triggers into models to induce malicious outcomes. Existing backdoor attacks targeting DNN-based TSC models remain elementary. In particular, early methods borrow trigger designs from computer vision, which are ineffective for time series data. More recent approaches utilize generative models for trigger generation, but at the cost of significant computational complexity. In this work, we analyze the limitations of existing attacks and introduce an enhanced method, FreqBack. Drawing inspiration from the fact that DNN models inherently capture frequency domain features in time series data, we identify that improper perturbations in the frequency domain are the root cause of ineffective attacks. To address this, we propose to generate triggers both effectively and efficiently, guided by frequency analysis. FreqBack exhibits substantial performance across five models and eight datasets, achieving an impressive attack success rate of over 90%, while maintaining less than a 3% drop in model accuracy on clean data.
How Well Do Supervised 3D Models Transfer to Medical Imaging Tasks?
Jan 20, 2025



Abstract:The pre-training and fine-tuning paradigm has become prominent in transfer learning. For example, if the model is pre-trained on ImageNet and then fine-tuned to PASCAL, it can significantly outperform that trained on PASCAL from scratch. While ImageNet pre-training has shown enormous success, it is formed in 2D, and the learned features are for classification tasks; when transferring to more diverse tasks, like 3D image segmentation, its performance is inevitably compromised due to the deviation from the original ImageNet context. A significant challenge lies in the lack of large, annotated 3D datasets rivaling the scale of ImageNet for model pre-training. To overcome this challenge, we make two contributions. Firstly, we construct AbdomenAtlas 1.1 that comprises 9,262 three-dimensional computed tomography (CT) volumes with high-quality, per-voxel annotations of 25 anatomical structures and pseudo annotations of seven tumor types. Secondly, we develop a suite of models that are pre-trained on our AbdomenAtlas 1.1 for transfer learning. Our preliminary analyses indicate that the model trained only with 21 CT volumes, 672 masks, and 40 GPU hours has a transfer learning ability similar to the model trained with 5,050 (unlabeled) CT volumes and 1,152 GPU hours. More importantly, the transfer learning ability of supervised models can further scale up with larger annotated datasets, achieving significantly better performance than preexisting pre-trained models, irrespective of their pre-training methodologies or data sources. We hope this study can facilitate collective efforts in constructing larger 3D medical datasets and more releases of supervised pre-trained models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge