Jieneng Chen
CausalSpatial: A Benchmark for Object-Centric Causal Spatial Reasoning
Jan 19, 2026Abstract:Humans can look at a static scene and instantly predict what happens next -- will moving this object cause a collision? We call this ability Causal Spatial Reasoning. However, current multimodal large language models (MLLMs) cannot do this, as they remain largely restricted to static spatial perception, struggling to answer "what-if" questions in a 3D scene. We introduce CausalSpatial, a diagnostic benchmark evaluating whether models can anticipate consequences of object motions across four tasks: Collision, Compatibility, Occlusion, and Trajectory. Results expose a severe gap: humans score 84% while GPT-5 achieves only 54%. Why do MLLMs fail? Our analysis uncovers a fundamental deficiency: models over-rely on textual chain-of-thought reasoning that drifts from visual evidence, producing fluent but spatially ungrounded hallucinations. To address this, we propose the Causal Object World model (COW), a framework that externalizes the simulation process by generating videos of hypothetical dynamics. With explicit visual cues of causality, COW enables models to ground their reasoning in physical reality rather than linguistic priors. We make the dataset and code publicly available here: https://github.com/CausalSpatial/CausalSpatial
Vision-Language-Vision Auto-Encoder: Scalable Knowledge Distillation from Diffusion Models
Jul 09, 2025Abstract:Building state-of-the-art Vision-Language Models (VLMs) with strong captioning capabilities typically necessitates training on billions of high-quality image-text pairs, requiring millions of GPU hours. This paper introduces the Vision-Language-Vision (VLV) auto-encoder framework, which strategically leverages key pretrained components: a vision encoder, the decoder of a Text-to-Image (T2I) diffusion model, and subsequently, a Large Language Model (LLM). Specifically, we establish an information bottleneck by regularizing the language representation space, achieved through freezing the pretrained T2I diffusion decoder. Our VLV pipeline effectively distills knowledge from the text-conditioned diffusion model using continuous embeddings, demonstrating comprehensive semantic understanding via high-quality reconstructions. Furthermore, by fine-tuning a pretrained LLM to decode the intermediate language representations into detailed descriptions, we construct a state-of-the-art (SoTA) captioner comparable to leading models like GPT-4o and Gemini 2.0 Flash. Our method demonstrates exceptional cost-efficiency and significantly reduces data requirements; by primarily utilizing single-modal images for training and maximizing the utility of existing pretrained models (image encoder, T2I diffusion model, and LLM), it circumvents the need for massive paired image-text datasets, keeping the total training expenditure under $1,000 USD.
SpatialLLM: A Compound 3D-Informed Design towards Spatially-Intelligent Large Multimodal Models
May 01, 2025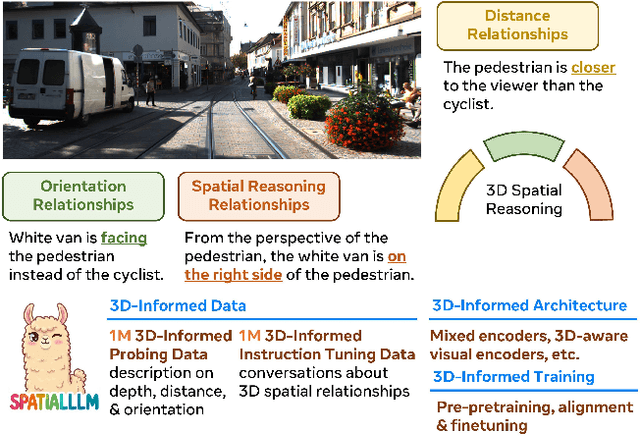
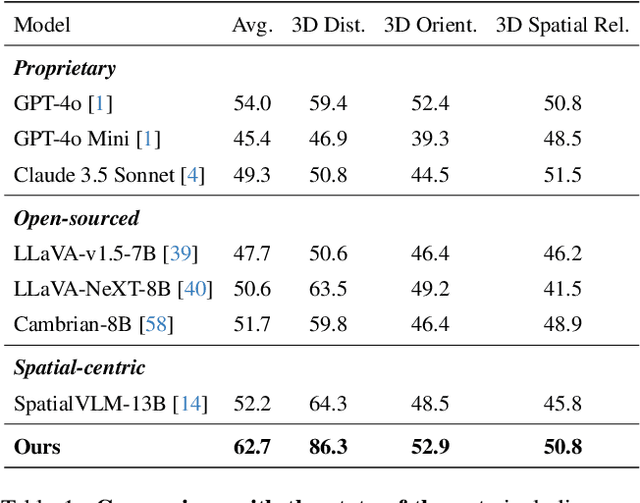
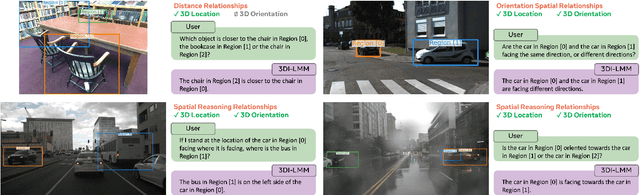
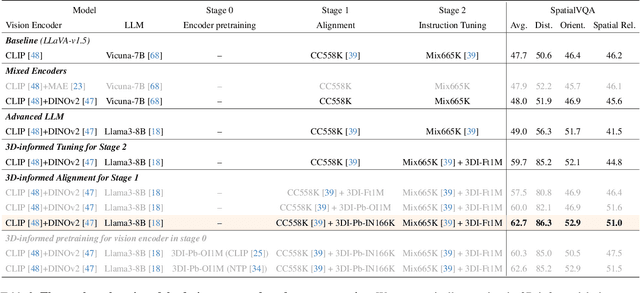
Abstract:Humans naturally understand 3D spatial relationships, enabling complex reasoning like predicting collisions of vehicles from different directions. Current large multimodal models (LMMs), however, lack of this capability of 3D spatial reasoning. This limitation stems from the scarcity of 3D training data and the bias in current model designs toward 2D data. In this paper, we systematically study the impact of 3D-informed data, architecture, and training setups, introducing SpatialLLM, a large multi-modal model with advanced 3D spatial reasoning abilities. To address data limitations, we develop two types of 3D-informed training datasets: (1) 3D-informed probing data focused on object's 3D location and orientation, and (2) 3D-informed conversation data for complex spatial relationships. Notably, we are the first to curate VQA data that incorporate 3D orientation relationships on real images. Furthermore, we systematically integrate these two types of training data with the architectural and training designs of LMMs, providing a roadmap for optimal design aimed at achieving superior 3D reasoning capabilities. Our SpatialLLM advances machines toward highly capable 3D-informed reasoning, surpassing GPT-4o performance by 8.7%. Our systematic empirical design and the resulting findings offer valuable insights for future research in this direction.
SpatialReasoner: Towards Explicit and Generalizable 3D Spatial Reasoning
Apr 28, 2025



Abstract:Recent studies in 3D spatial reasoning explore data-driven approaches and achieve enhanced spatial reasoning performance with reinforcement learning (RL). However, these methods typically perform spatial reasoning in an implicit manner, and it remains underexplored whether the acquired 3D knowledge generalizes to unseen question types at any stage of the training. In this work we introduce SpatialReasoner, a novel large vision-language model (LVLM) that address 3D spatial reasoning with explicit 3D representations shared between stages -- 3D perception, computation, and reasoning. Explicit 3D representations provide a coherent interface that supports advanced 3D spatial reasoning and enable us to study the factual errors made by LVLMs. Results show that our SpatialReasoner achieve improved performance on a variety of spatial reasoning benchmarks and generalizes better when evaluating on novel 3D spatial reasoning questions. Our study bridges the 3D parsing capabilities of prior visual foundation models with the powerful reasoning abilities of large language models, opening new directions for 3D spatial reasoning.
PulseCheck457: A Diagnostic Benchmark for 6D Spatial Reasoning of Large Multimodal Models
Feb 13, 2025Abstract:Although large multimodal models (LMMs) have demonstrated remarkable capabilities in visual scene interpretation and reasoning, their capacity for complex and precise 3-dimensional spatial reasoning remains uncertain. Existing benchmarks focus predominantly on 2D spatial understanding and lack a framework to comprehensively evaluate 6D spatial reasoning across varying complexities. To address this limitation, we present PulseCheck457, a scalable and unbiased synthetic dataset designed with 4 key capability for spatial reasoning: multi-object recognition, 2D location, 3D location, and 3D orientation. We develop a cascading evaluation structure, constructing 7 question types across 5 difficulty levels that range from basic single object recognition to our new proposed complex 6D spatial reasoning tasks. We evaluated various large multimodal models (LMMs) on PulseCheck457, observing a general decline in performance as task complexity increases, particularly in 3D reasoning and 6D spatial tasks. To quantify these challenges, we introduce the Relative Performance Dropping Rate (RPDR), highlighting key weaknesses in 3D reasoning capabilities. Leveraging the unbiased attribute design of our dataset, we also uncover prediction biases across different attributes, with similar patterns observed in real-world image settings.
PulseCheck457: A Diagnostic Benchmark for Comprehensive Spatial Reasoning of Large Multimodal Models
Feb 12, 2025Abstract:Although large multimodal models (LMMs) have demonstrated remarkable capabilities in visual scene interpretation and reasoning, their capacity for complex and precise 3-dimensional spatial reasoning remains uncertain. Existing benchmarks focus predominantly on 2D spatial understanding and lack a framework to comprehensively evaluate 6D spatial reasoning across varying complexities. To address this limitation, we present PulseCheck457, a scalable and unbiased synthetic dataset designed with 4 key capability for spatial reasoning: multi-object recognition, 2D location, 3D location, and 3D orientation. We develop a cascading evaluation structure, constructing 7 question types across 5 difficulty levels that range from basic single object recognition to our new proposed complex 6D spatial reasoning tasks. We evaluated various large multimodal models (LMMs) on PulseCheck457, observing a general decline in performance as task complexity increases, particularly in 3D reasoning and 6D spatial tasks. To quantify these challenges, we introduce the Relative Performance Dropping Rate (RPDR), highlighting key weaknesses in 3D reasoning capabilities. Leveraging the unbiased attribute design of our dataset, we also uncover prediction biases across different attributes, with similar patterns observed in real-world image settings.
GenEx: Generating an Explorable World
Dec 12, 2024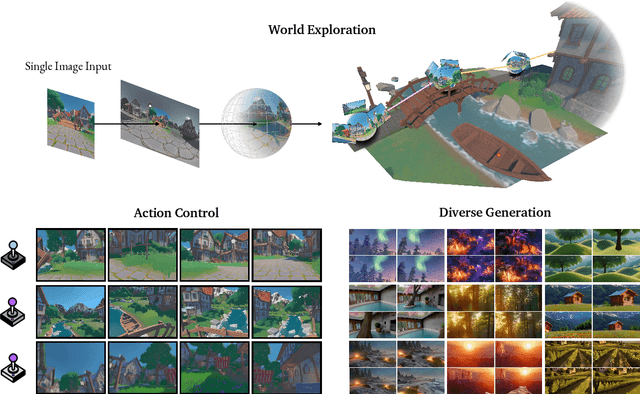
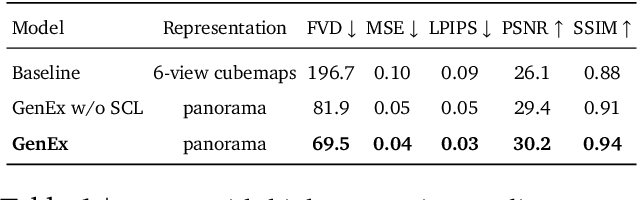

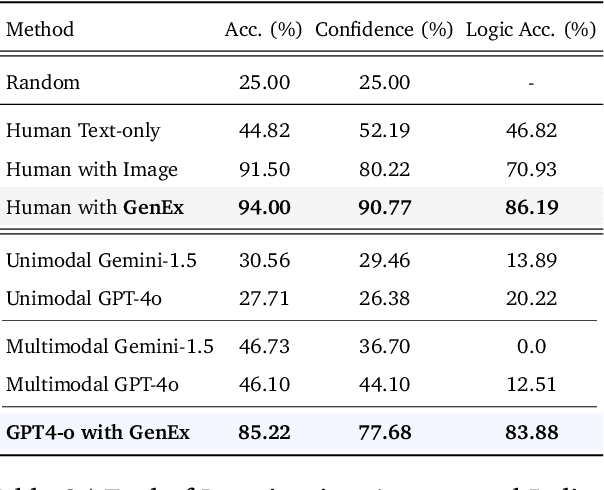
Abstract:Understanding, navigating, and exploring the 3D physical real world has long been a central challenge in the development of artificial intelligence. In this work, we take a step toward this goal by introducing GenEx, a system capable of planning complex embodied world exploration, guided by its generative imagination that forms priors (expectations) about the surrounding environments. GenEx generates an entire 3D-consistent imaginative environment from as little as a single RGB image, bringing it to life through panoramic video streams. Leveraging scalable 3D world data curated from Unreal Engine, our generative model is rounded in the physical world. It captures a continuous 360-degree environment with little effort, offering a boundless landscape for AI agents to explore and interact with. GenEx achieves high-quality world generation, robust loop consistency over long trajectories, and demonstrates strong 3D capabilities such as consistency and active 3D mapping. Powered by generative imagination of the world, GPT-assisted agents are equipped to perform complex embodied tasks, including both goal-agnostic exploration and goal-driven navigation. These agents utilize predictive expectation regarding unseen parts of the physical world to refine their beliefs, simulate different outcomes based on potential decisions, and make more informed choices. In summary, we demonstrate that GenEx provides a transformative platform for advancing embodied AI in imaginative spaces and brings potential for extending these capabilities to real-world exploration.
3DSRBench: A Comprehensive 3D Spatial Reasoning Benchmark
Dec 10, 2024Abstract:3D spatial reasoning is the ability to analyze and interpret the positions, orientations, and spatial relationships of objects within the 3D space. This allows models to develop a comprehensive understanding of the 3D scene, enabling their applicability to a broader range of areas, such as autonomous navigation, robotics, and AR/VR. While large multi-modal models (LMMs) have achieved remarkable progress in a wide range of image and video understanding tasks, their capabilities to perform 3D spatial reasoning on diverse natural images are less studied. In this work we present the first comprehensive 3D spatial reasoning benchmark, 3DSRBench, with 2,772 manually annotated visual question-answer pairs across 12 question types. We conduct robust and thorough evaluation of 3D spatial reasoning capabilities by balancing the data distribution and adopting a novel FlipEval strategy. To further study the robustness of 3D spatial reasoning w.r.t. camera 3D viewpoints, our 3DSRBench includes two subsets with 3D spatial reasoning questions on paired images with common and uncommon viewpoints. We benchmark a wide range of open-sourced and proprietary LMMs, uncovering their limitations in various aspects of 3D awareness, such as height, orientation, location, and multi-object reasoning, as well as their degraded performance on images with uncommon camera viewpoints. Our 3DSRBench provide valuable findings and insights about the future development of LMMs with strong 3D reasoning capabilities. Our project page and dataset is available https://3dsrbench.github.io.
Generative World Explorer
Nov 19, 2024



Abstract:Planning with partial observation is a central challenge in embodied AI. A majority of prior works have tackled this challenge by developing agents that physically explore their environment to update their beliefs about the world state. In contrast, humans can $\textit{imagine}$ unseen parts of the world through a mental exploration and $\textit{revise}$ their beliefs with imagined observations. Such updated beliefs can allow them to make more informed decisions, without necessitating the physical exploration of the world at all times. To achieve this human-like ability, we introduce the $\textit{Generative World Explorer (Genex)}$, an egocentric world exploration framework that allows an agent to mentally explore a large-scale 3D world (e.g., urban scenes) and acquire imagined observations to update its belief. This updated belief will then help the agent to make a more informed decision at the current step. To train $\textit{Genex}$, we create a synthetic urban scene dataset, Genex-DB. Our experimental results demonstrate that (1) $\textit{Genex}$ can generate high-quality and consistent observations during long-horizon exploration of a large virtual physical world and (2) the beliefs updated with the generated observations can inform an existing decision-making model (e.g., an LLM agent) to make better plans.
Touchstone Benchmark: Are We on the Right Way for Evaluating AI Algorithms for Medical Segmentation?
Nov 06, 2024



Abstract:How can we test AI performance? This question seems trivial, but it isn't. Standard benchmarks often have problems such as in-distribution and small-size test sets, oversimplified metrics, unfair comparisons, and short-term outcome pressure. As a consequence, good performance on standard benchmarks does not guarantee success in real-world scenarios. To address these problems, we present Touchstone, a large-scale collaborative segmentation benchmark of 9 types of abdominal organs. This benchmark is based on 5,195 training CT scans from 76 hospitals around the world and 5,903 testing CT scans from 11 additional hospitals. This diverse test set enhances the statistical significance of benchmark results and rigorously evaluates AI algorithms across various out-of-distribution scenarios. We invited 14 inventors of 19 AI algorithms to train their algorithms, while our team, as a third party, independently evaluated these algorithms on three test sets. In addition, we also evaluated pre-existing AI frameworks--which, differing from algorithms, are more flexible and can support different algorithms--including MONAI from NVIDIA, nnU-Net from DKFZ, and numerous other open-source frameworks. We are committed to expanding this benchmark to encourage more innovation of AI algorithms for the medical domain.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge