Yu-Cheng Chou
Generative Adversarial Reasoner: Enhancing LLM Reasoning with Adversarial Reinforcement Learning
Dec 18, 2025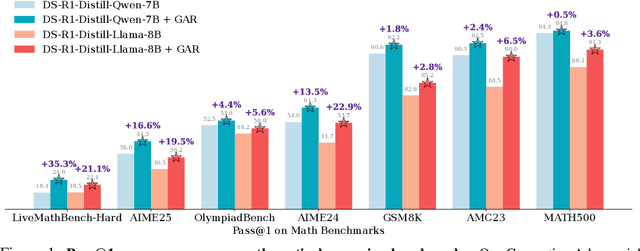
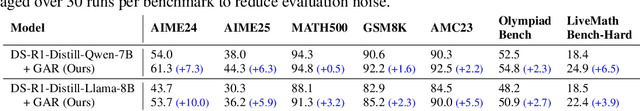
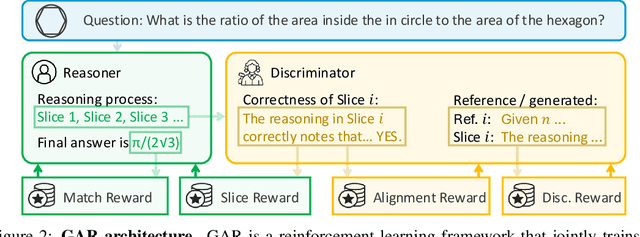
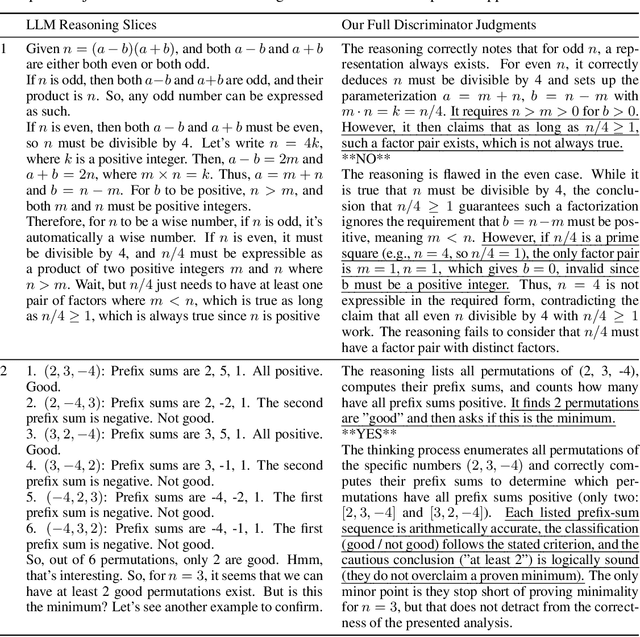
Abstract:Large language models (LLMs) with explicit reasoning capabilities excel at mathematical reasoning yet still commit process errors, such as incorrect calculations, brittle logic, and superficially plausible but invalid steps. In this paper, we introduce Generative Adversarial Reasoner, an on-policy joint training framework designed to enhance reasoning by co-evolving an LLM reasoner and an LLM-based discriminator through adversarial reinforcement learning. A compute-efficient review schedule partitions each reasoning chain into logically complete slices of comparable length, and the discriminator evaluates each slice's soundness with concise, structured justifications. Learning couples complementary signals: the LLM reasoner is rewarded for logically consistent steps that yield correct answers, while the discriminator earns rewards for correctly detecting errors or distinguishing traces in the reasoning process. This produces dense, well-calibrated, on-policy step-level rewards that supplement sparse exact-match signals, improving credit assignment, increasing sample efficiency, and enhancing overall reasoning quality of LLMs. Across various mathematical benchmarks, the method delivers consistent gains over strong baselines with standard RL post-training. Specifically, on AIME24, we improve DeepSeek-R1-Distill-Qwen-7B from 54.0 to 61.3 (+7.3) and DeepSeek-R1-Distill-Llama-8B from 43.7 to 53.7 (+10.0). The modular discriminator also enables flexible reward shaping for objectives such as teacher distillation, preference alignment, and mathematical proof-based reasoning.
SpatialReasoner: Towards Explicit and Generalizable 3D Spatial Reasoning
Apr 28, 2025



Abstract:Recent studies in 3D spatial reasoning explore data-driven approaches and achieve enhanced spatial reasoning performance with reinforcement learning (RL). However, these methods typically perform spatial reasoning in an implicit manner, and it remains underexplored whether the acquired 3D knowledge generalizes to unseen question types at any stage of the training. In this work we introduce SpatialReasoner, a novel large vision-language model (LVLM) that address 3D spatial reasoning with explicit 3D representations shared between stages -- 3D perception, computation, and reasoning. Explicit 3D representations provide a coherent interface that supports advanced 3D spatial reasoning and enable us to study the factual errors made by LVLMs. Results show that our SpatialReasoner achieve improved performance on a variety of spatial reasoning benchmarks and generalizes better when evaluating on novel 3D spatial reasoning questions. Our study bridges the 3D parsing capabilities of prior visual foundation models with the powerful reasoning abilities of large language models, opening new directions for 3D spatial reasoning.
Ultrasound Image Synthesis Using Generative AI for Lung Ultrasound Detection
Jan 10, 2025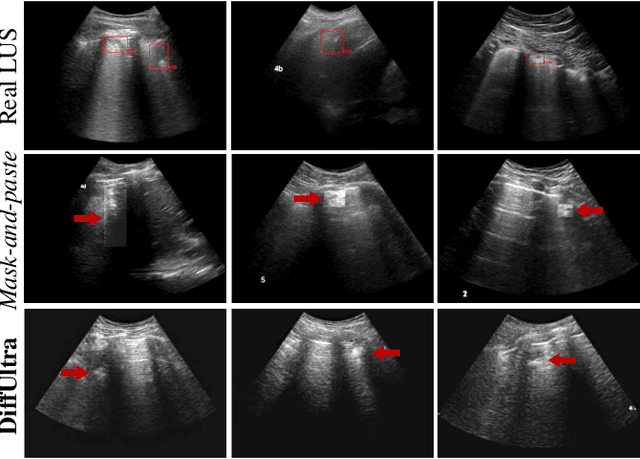

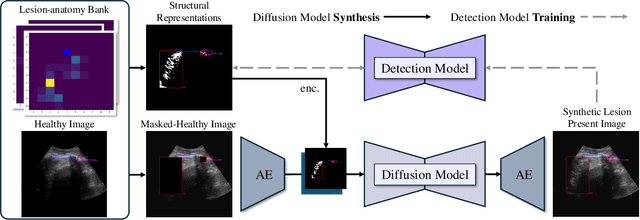

Abstract:Developing reliable healthcare AI models requires training with representative and diverse data. In imbalanced datasets, model performance tends to plateau on the more prevalent classes while remaining low on less common cases. To overcome this limitation, we propose DiffUltra, the first generative AI technique capable of synthesizing realistic Lung Ultrasound (LUS) images with extensive lesion variability. Specifically, we condition the generative AI by the introduced Lesion-anatomy Bank, which captures the lesion's structural and positional properties from real patient data to guide the image synthesis.We demonstrate that DiffUltra improves consolidation detection by 5.6% in AP compared to the models trained solely on real patient data. More importantly, DiffUltra increases data diversity and prevalence of rare cases, leading to a 25% AP improvement in detecting rare instances such as large lung consolidations, which make up only 10% of the dataset.
ScaleMAI: Accelerating the Development of Trusted Datasets and AI Models
Jan 06, 2025

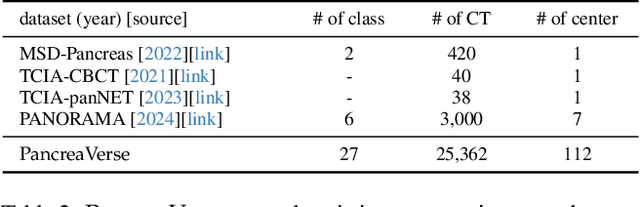

Abstract:Building trusted datasets is critical for transparent and responsible Medical AI (MAI) research, but creating even small, high-quality datasets can take years of effort from multidisciplinary teams. This process often delays AI benefits, as human-centric data creation and AI-centric model development are treated as separate, sequential steps. To overcome this, we propose ScaleMAI, an agent of AI-integrated data curation and annotation, allowing data quality and AI performance to improve in a self-reinforcing cycle and reducing development time from years to months. We adopt pancreatic tumor detection as an example. First, ScaleMAI progressively creates a dataset of 25,362 CT scans, including per-voxel annotations for benign/malignant tumors and 24 anatomical structures. Second, through progressive human-in-the-loop iterations, ScaleMAI provides Flagship AI Model that can approach the proficiency of expert annotators (30-year experience) in detecting pancreatic tumors. Flagship Model significantly outperforms models developed from smaller, fixed-quality datasets, with substantial gains in tumor detection (+14%), segmentation (+5%), and classification (72%) on three prestigious benchmarks. In summary, ScaleMAI transforms the speed, scale, and reliability of medical dataset creation, paving the way for a variety of impactful, data-driven applications.
Touchstone Benchmark: Are We on the Right Way for Evaluating AI Algorithms for Medical Segmentation?
Nov 06, 2024



Abstract:How can we test AI performance? This question seems trivial, but it isn't. Standard benchmarks often have problems such as in-distribution and small-size test sets, oversimplified metrics, unfair comparisons, and short-term outcome pressure. As a consequence, good performance on standard benchmarks does not guarantee success in real-world scenarios. To address these problems, we present Touchstone, a large-scale collaborative segmentation benchmark of 9 types of abdominal organs. This benchmark is based on 5,195 training CT scans from 76 hospitals around the world and 5,903 testing CT scans from 11 additional hospitals. This diverse test set enhances the statistical significance of benchmark results and rigorously evaluates AI algorithms across various out-of-distribution scenarios. We invited 14 inventors of 19 AI algorithms to train their algorithms, while our team, as a third party, independently evaluated these algorithms on three test sets. In addition, we also evaluated pre-existing AI frameworks--which, differing from algorithms, are more flexible and can support different algorithms--including MONAI from NVIDIA, nnU-Net from DKFZ, and numerous other open-source frameworks. We are committed to expanding this benchmark to encourage more innovation of AI algorithms for the medical domain.
Embracing Massive Medical Data
Jul 05, 2024Abstract:As massive medical data become available with an increasing number of scans, expanding classes, and varying sources, prevalent training paradigms -- where AI is trained with multiple passes over fixed, finite datasets -- face significant challenges. First, training AI all at once on such massive data is impractical as new scans/sources/classes continuously arrive. Second, training AI continuously on new scans/sources/classes can lead to catastrophic forgetting, where AI forgets old data as it learns new data, and vice versa. To address these two challenges, we propose an online learning method that enables training AI from massive medical data. Instead of repeatedly training AI on randomly selected data samples, our method identifies the most significant samples for the current AI model based on their data uniqueness and prediction uncertainty, then trains the AI on these selective data samples. Compared with prevalent training paradigms, our method not only improves data efficiency by enabling training on continual data streams, but also mitigates catastrophic forgetting by selectively training AI on significant data samples that might otherwise be forgotten, outperforming by 15% in Dice score for multi-organ and tumor segmentation. The code is available at https://github.com/MrGiovanni/OnlineLearning
Acquiring Weak Annotations for Tumor Localization in Temporal and Volumetric Data
Oct 23, 2023Abstract:Creating large-scale and well-annotated datasets to train AI algorithms is crucial for automated tumor detection and localization. However, with limited resources, it is challenging to determine the best type of annotations when annotating massive amounts of unlabeled data. To address this issue, we focus on polyps in colonoscopy videos and pancreatic tumors in abdominal CT scans; both applications require significant effort and time for pixel-wise annotation due to the high dimensional nature of the data, involving either temporary or spatial dimensions. In this paper, we develop a new annotation strategy, termed Drag&Drop, which simplifies the annotation process to drag and drop. This annotation strategy is more efficient, particularly for temporal and volumetric imaging, than other types of weak annotations, such as per-pixel, bounding boxes, scribbles, ellipses, and points. Furthermore, to exploit our Drag&Drop annotations, we develop a novel weakly supervised learning method based on the watershed algorithm. Experimental results show that our method achieves better detection and localization performance than alternative weak annotations and, more importantly, achieves similar performance to that trained on detailed per-pixel annotations. Interestingly, we find that, with limited resources, allocating weak annotations from a diverse patient population can foster models more robust to unseen images than allocating per-pixel annotations for a small set of images. In summary, this research proposes an efficient annotation strategy for tumor detection and localization that is less accurate than per-pixel annotations but useful for creating large-scale datasets for screening tumors in various medical modalities.
Early Detection and Localization of Pancreatic Cancer by Label-Free Tumor Synthesis
Aug 06, 2023



Abstract:Early detection and localization of pancreatic cancer can increase the 5-year survival rate for patients from 8.5% to 20%. Artificial intelligence (AI) can potentially assist radiologists in detecting pancreatic tumors at an early stage. Training AI models require a vast number of annotated examples, but the availability of CT scans obtaining early-stage tumors is constrained. This is because early-stage tumors may not cause any symptoms, which can delay detection, and the tumors are relatively small and may be almost invisible to human eyes on CT scans. To address this issue, we develop a tumor synthesis method that can synthesize enormous examples of small pancreatic tumors in the healthy pancreas without the need for manual annotation. Our experiments demonstrate that the overall detection rate of pancreatic tumors, measured by Sensitivity and Specificity, achieved by AI trained on synthetic tumors is comparable to that of real tumors. More importantly, our method shows a much higher detection rate for small tumors. We further investigate the per-voxel segmentation performance of pancreatic tumors if AI is trained on a combination of CT scans with synthetic tumors and CT scans with annotated large tumors at an advanced stage. Finally, we show that synthetic tumors improve AI generalizability in tumor detection and localization when processing CT scans from different hospitals. Overall, our proposed tumor synthesis method has immense potential to improve the early detection of pancreatic cancer, leading to better patient outcomes.
Deep Gradient Learning for Efficient Camouflaged Object Detection
May 25, 2022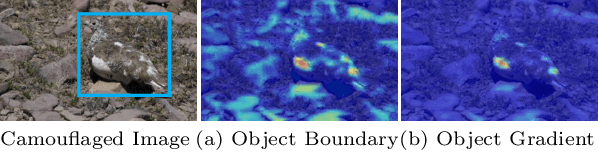
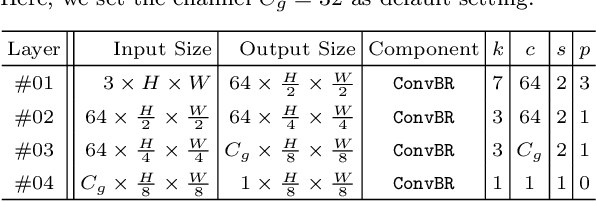
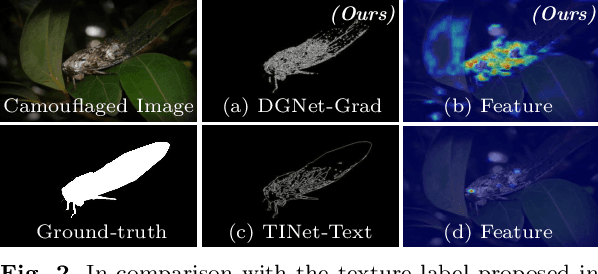
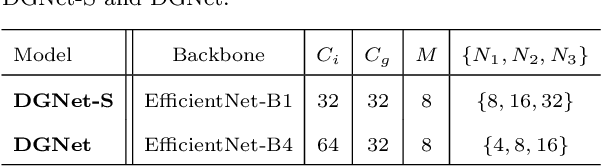
Abstract:This paper introduces DGNet, a novel deep framework that exploits object gradient supervision for camouflaged object detection (COD). It decouples the task into two connected branches, i.e., a context and a texture encoder. The essential connection is the gradient-induced transition, representing a soft grouping between context and texture features. Benefiting from the simple but efficient framework, DGNet outperforms existing state-of-the-art COD models by a large margin. Notably, our efficient version, DGNet-S, runs in real-time (80 fps) and achieves comparable results to the cutting-edge model JCSOD-CVPR$_{21}$ with only 6.82% parameters. Application results also show that the proposed DGNet performs well in polyp segmentation, defect detection, and transparent object segmentation tasks. Codes will be made available at https://github.com/GewelsJI/DGNet.
Video Polyp Segmentation: A Deep Learning Perspective
Mar 27, 2022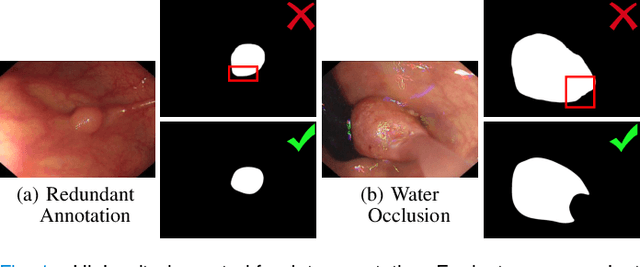
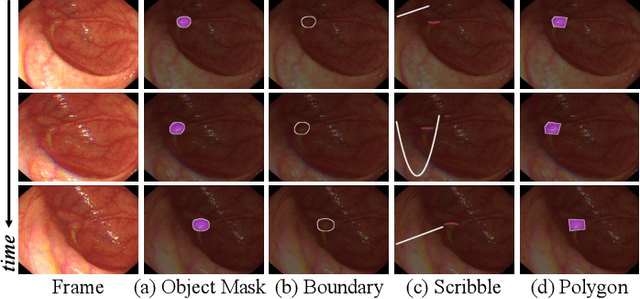
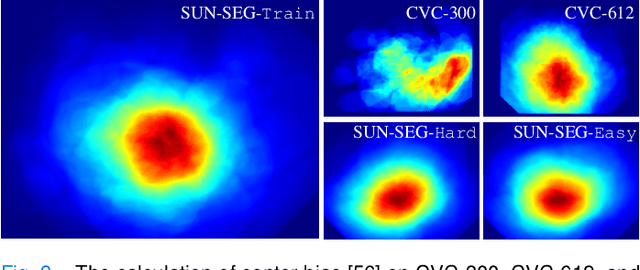
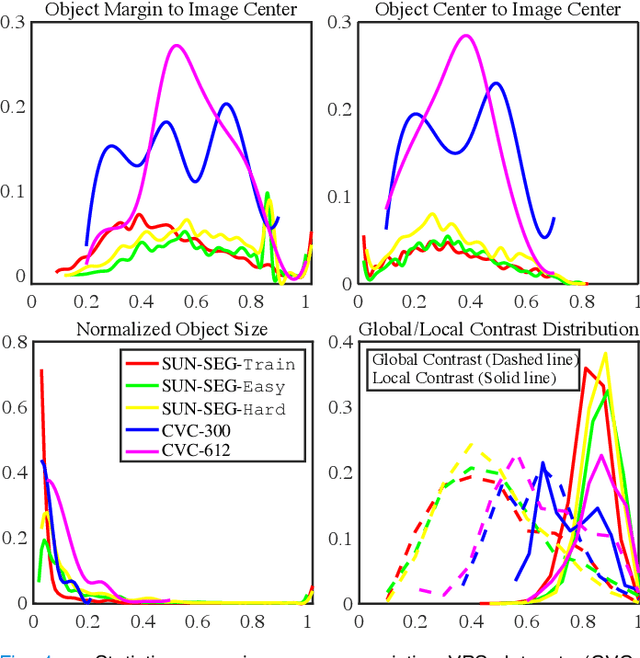
Abstract:In the deep learning era, we present the first comprehensive video polyp segmentation (VPS) study. Over the years, developments in VPS are not moving forward with ease due to the lack of large-scale fine-grained segmentation annotations. To tackle this issue, we first introduce a high-quality per-frame annotated VPS dataset, named SUN-SEG, which includes 158,690 frames from the famous SUN dataset. We provide additional annotations with diverse types, i.e., attribute, object mask, boundary, scribble, and polygon. Second, we design a simple but efficient baseline, dubbed PNS+, consisting of a global encoder, a local encoder, and normalized self-attention (NS) blocks. The global and local encoders receive an anchor frame and multiple successive frames to extract long-term and short-term feature representations, which are then progressively updated by two NS blocks. Extensive experiments show that PNS+ achieves the best performance and real-time inference speed (170fps), making it a promising solution for the VPS task. Third, we extensively evaluate 13 representative polyp/object segmentation models on our SUN-SEG dataset and provide attribute-based comparisons. Benchmark results are available at https: //github.com/GewelsJI/VPS.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge