Shuwen Sun
Interpretable Spatial-Temporal Fusion Transformers: Multi-Output Prediction for Parametric Dynamical Systems with Time-Varying Inputs
May 01, 2025


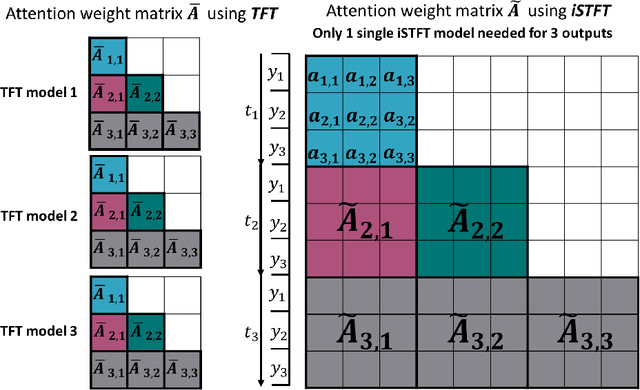
Abstract:We explore the promising performance of a transformer model in predicting outputs of parametric dynamical systems with external time-varying input signals. The outputs of such systems vary not only with physical parameters but also with external time-varying input signals. Accurately catching the dynamics of such systems is challenging. We have adapted and extended an existing transformer model for single output prediction to a multiple-output transformer that is able to predict multiple output responses of these systems. The multiple-output transformer generalizes the interpretability of the original transformer. The generalized interpretable attention weight matrix explores not only the temporal correlations in the sequence, but also the interactions between the multiple outputs, providing explanation for the spatial correlation in the output domain. This multiple-output transformer accurately predicts the sequence of multiple outputs, regardless of the nonlinearity of the system and the dimensionality of the parameter space.
Data-Augmented Predictive Deep Neural Network: Enhancing the extrapolation capabilities of non-intrusive surrogate models
Oct 17, 2024



Abstract:Numerically solving a large parametric nonlinear dynamical system is challenging due to its high complexity and the high computational costs. In recent years, machine-learning-aided surrogates are being actively researched. However, many methods fail in accurately generalizing in the entire time interval $[0, T]$, when the training data is available only in a training time interval $[0, T_0]$, with $T_0<T$. To improve the extrapolation capabilities of the surrogate models in the entire time domain, we propose a new deep learning framework, where kernel dynamic mode decomposition (KDMD) is employed to evolve the dynamics of the latent space generated by the encoder part of a convolutional autoencoder (CAE). After adding the KDMD-decoder-extrapolated data into the original data set, we train the CAE along with a feed-forward deep neural network using the augmented data. The trained network can predict future states outside the training time interval at any out-of-training parameter samples. The proposed method is tested on two numerical examples: a FitzHugh-Nagumo model and a model of incompressible flow past a cylinder. Numerical results show accurate and fast prediction performance in both the time and the parameter domain.
Early Detection and Localization of Pancreatic Cancer by Label-Free Tumor Synthesis
Aug 06, 2023



Abstract:Early detection and localization of pancreatic cancer can increase the 5-year survival rate for patients from 8.5% to 20%. Artificial intelligence (AI) can potentially assist radiologists in detecting pancreatic tumors at an early stage. Training AI models require a vast number of annotated examples, but the availability of CT scans obtaining early-stage tumors is constrained. This is because early-stage tumors may not cause any symptoms, which can delay detection, and the tumors are relatively small and may be almost invisible to human eyes on CT scans. To address this issue, we develop a tumor synthesis method that can synthesize enormous examples of small pancreatic tumors in the healthy pancreas without the need for manual annotation. Our experiments demonstrate that the overall detection rate of pancreatic tumors, measured by Sensitivity and Specificity, achieved by AI trained on synthetic tumors is comparable to that of real tumors. More importantly, our method shows a much higher detection rate for small tumors. We further investigate the per-voxel segmentation performance of pancreatic tumors if AI is trained on a combination of CT scans with synthetic tumors and CT scans with annotated large tumors at an advanced stage. Finally, we show that synthetic tumors improve AI generalizability in tumor detection and localization when processing CT scans from different hospitals. Overall, our proposed tumor synthesis method has immense potential to improve the early detection of pancreatic cancer, leading to better patient outcomes.
Label-Free Liver Tumor Segmentation
Mar 27, 2023
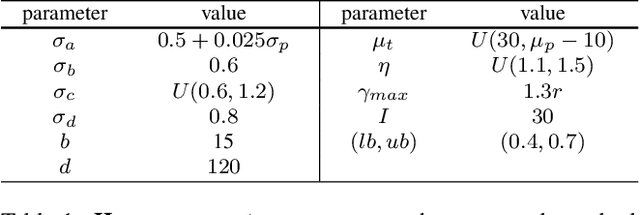

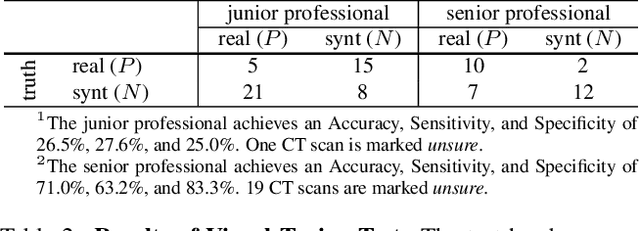
Abstract:We demonstrate that AI models can accurately segment liver tumors without the need for manual annotation by using synthetic tumors in CT scans. Our synthetic tumors have two intriguing advantages: (I) realistic in shape and texture, which even medical professionals can confuse with real tumors; (II) effective for training AI models, which can perform liver tumor segmentation similarly to the model trained on real tumors -- this result is exciting because no existing work, using synthetic tumors only, has thus far reached a similar or even close performance to real tumors. This result also implies that manual efforts for annotating tumors voxel by voxel (which took years to create) can be significantly reduced in the future. Moreover, our synthetic tumors can automatically generate many examples of small (or even tiny) synthetic tumors and have the potential to improve the success rate of detecting small liver tumors, which is critical for detecting the early stages of cancer. In addition to enriching the training data, our synthesizing strategy also enables us to rigorously assess the AI robustness.
Synthetic Tumors Make AI Segment Tumors Better
Oct 26, 2022

Abstract:We develop a novel strategy to generate synthetic tumors. Unlike existing works, the tumors generated by our strategy have two intriguing advantages: (1) realistic in shape and texture, which even medical professionals can confuse with real tumors; (2) effective for AI model training, which can perform liver tumor segmentation similarly to a model trained on real tumors - this result is unprecedented because no existing work, using synthetic tumors only, has thus far reached a similar or even close performance to the model trained on real tumors. This result also implies that manual efforts for developing per-voxel annotation of tumors (which took years to create) can be considerably reduced for training AI models in the future. Moreover, our synthetic tumors have the potential to improve the success rate of small tumor detection by automatically generating enormous examples of small (or tiny) synthetic tumors.
Sequential Learning on Liver Tumor Boundary Semantics and Prognostic Biomarker Mining
Mar 09, 2021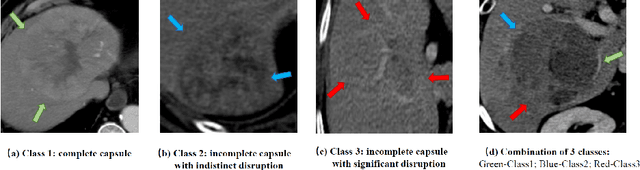
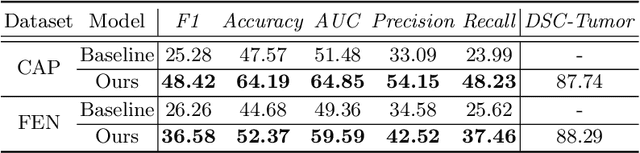
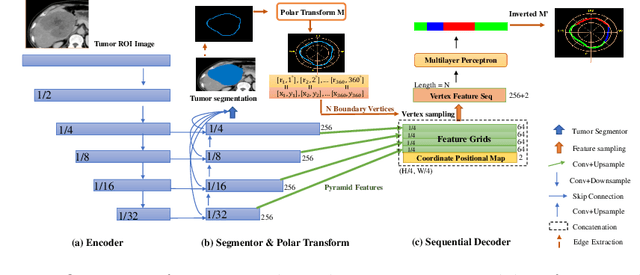
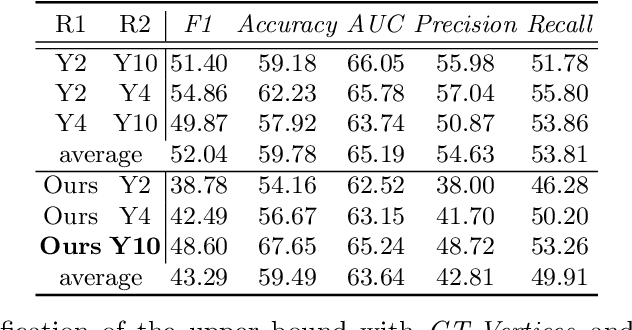
Abstract:The boundary of tumors (hepatocellular carcinoma, or HCC) contains rich semantics: capsular invasion, visibility, smoothness, folding and protuberance, etc. Capsular invasion on tumor boundary has proven to be clinically correlated with the prognostic indicator, microvascular invasion (MVI). Investigating tumor boundary semantics has tremendous clinical values. In this paper, we propose the first and novel computational framework that disentangles the task into two components: spatial vertex localization and sequential semantic classification. (1) A HCC tumor segmentor is built for tumor mask boundary extraction, followed by polar transform representing the boundary with radius and angle. Vertex generator is used to produce fixed-length boundary vertices where vertex features are sampled on the corresponding spatial locations. (2) The sampled deep vertex features with positional embedding are mapped into a sequential space and decoded by a multilayer perceptron (MLP) for semantic classification. Extensive experiments on tumor capsule semantics demonstrate the effectiveness of our framework. Mining the correlation between the boundary semantics and MVI status proves the feasibility to integrate this boundary semantics as a valid HCC prognostic biomarker.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge