Hualin Qiao
Early and Prediagnostic Detection of Pancreatic Cancer from Computed Tomography
Jan 29, 2026Abstract:Pancreatic ductal adenocarcinoma (PDAC), one of the deadliest solid malignancies, is often detected at a late and inoperable stage. Retrospective reviews of prediagnostic CT scans, when conducted by expert radiologists aware that the patient later developed PDAC, frequently reveal lesions that were previously overlooked. To help detecting these lesions earlier, we developed an automated system named ePAI (early Pancreatic cancer detection with Artificial Intelligence). It was trained on data from 1,598 patients from a single medical center. In the internal test involving 1,009 patients, ePAI achieved an area under the receiver operating characteristic curve (AUC) of 0.939-0.999, a sensitivity of 95.3%, and a specificity of 98.7% for detecting small PDAC less than 2 cm in diameter, precisely localizing PDAC as small as 2 mm. In an external test involving 7,158 patients across 6 centers, ePAI achieved an AUC of 0.918-0.945, a sensitivity of 91.5%, and a specificity of 88.0%, precisely localizing PDAC as small as 5 mm. Importantly, ePAI detected PDACs on prediagnostic CT scans obtained 3 to 36 months before clinical diagnosis that had originally been overlooked by radiologists. It successfully detected and localized PDACs in 75 of 159 patients, with a median lead time of 347 days before clinical diagnosis. Our multi-reader study showed that ePAI significantly outperformed 30 board-certified radiologists by 50.3% (P < 0.05) in sensitivity while maintaining a comparable specificity of 95.4% in detecting PDACs early and prediagnostic. These findings suggest its potential of ePAI as an assistive tool to improve early detection of pancreatic cancer.
Auditing Significance, Metric Choice, and Demographic Fairness in Medical AI Challenges
Dec 22, 2025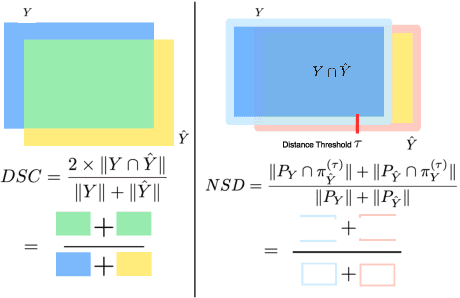
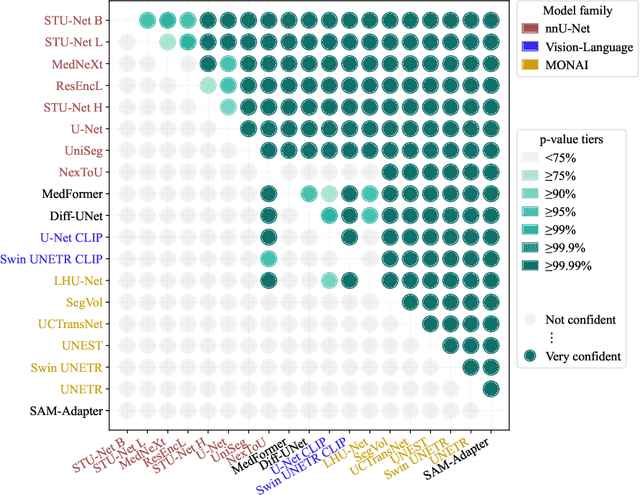
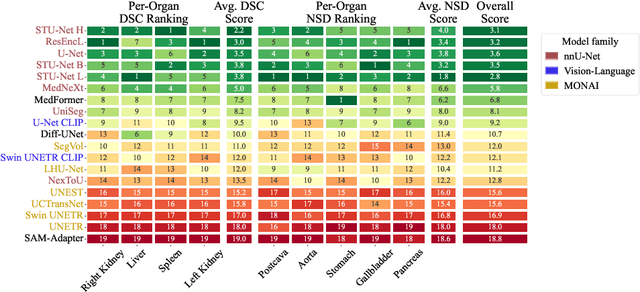
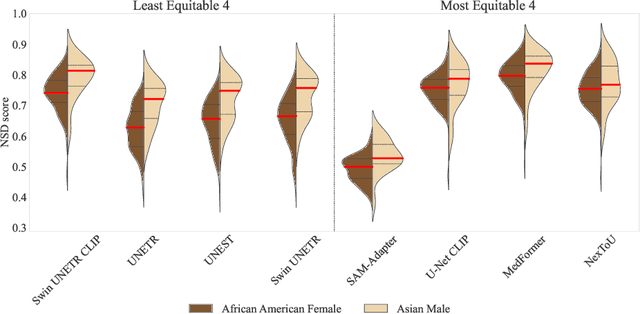
Abstract:Open challenges have become the de facto standard for comparative ranking of medical AI methods. Despite their importance, medical AI leaderboards exhibit three persistent limitations: (1) score gaps are rarely tested for statistical significance, so rank stability is unknown; (2) single averaged metrics are applied to every organ, hiding clinically important boundary errors; (3) performance across intersecting demographics is seldom reported, masking fairness and equity gaps. We introduce RankInsight, an open-source toolkit that seeks to address these limitations. RankInsight (1) computes pair-wise significance maps that show the nnU-Net family outperforms Vision-Language and MONAI submissions with high statistical certainty; (2) recomputes leaderboards with organ-appropriate metrics, reversing the order of the top four models when Dice is replaced by NSD for tubular structures; and (3) audits intersectional fairness, revealing that more than half of the MONAI-based entries have the largest gender-race discrepancy on our proprietary Johns Hopkins Hospital dataset. The RankInsight toolkit is publicly released and can be directly applied to past, ongoing, and future challenges. It enables organizers and participants to publish rankings that are statistically sound, clinically meaningful, and demographically fair.
Early Detection and Localization of Pancreatic Cancer by Label-Free Tumor Synthesis
Aug 06, 2023



Abstract:Early detection and localization of pancreatic cancer can increase the 5-year survival rate for patients from 8.5% to 20%. Artificial intelligence (AI) can potentially assist radiologists in detecting pancreatic tumors at an early stage. Training AI models require a vast number of annotated examples, but the availability of CT scans obtaining early-stage tumors is constrained. This is because early-stage tumors may not cause any symptoms, which can delay detection, and the tumors are relatively small and may be almost invisible to human eyes on CT scans. To address this issue, we develop a tumor synthesis method that can synthesize enormous examples of small pancreatic tumors in the healthy pancreas without the need for manual annotation. Our experiments demonstrate that the overall detection rate of pancreatic tumors, measured by Sensitivity and Specificity, achieved by AI trained on synthetic tumors is comparable to that of real tumors. More importantly, our method shows a much higher detection rate for small tumors. We further investigate the per-voxel segmentation performance of pancreatic tumors if AI is trained on a combination of CT scans with synthetic tumors and CT scans with annotated large tumors at an advanced stage. Finally, we show that synthetic tumors improve AI generalizability in tumor detection and localization when processing CT scans from different hospitals. Overall, our proposed tumor synthesis method has immense potential to improve the early detection of pancreatic cancer, leading to better patient outcomes.
Annotating 8,000 Abdominal CT Volumes for Multi-Organ Segmentation in Three Weeks
May 16, 2023Abstract:Annotating medical images, particularly for organ segmentation, is laborious and time-consuming. For example, annotating an abdominal organ requires an estimated rate of 30-60 minutes per CT volume based on the expertise of an annotator and the size, visibility, and complexity of the organ. Therefore, publicly available datasets for multi-organ segmentation are often limited in data size and organ diversity. This paper proposes a systematic and efficient method to expedite the annotation process for organ segmentation. We have created the largest multi-organ dataset (by far) with the spleen, liver, kidneys, stomach, gallbladder, pancreas, aorta, and IVC annotated in 8,448 CT volumes, equating to 3.2 million slices. The conventional annotation methods would take an experienced annotator up to 1,600 weeks (or roughly 30.8 years) to complete this task. In contrast, our annotation method has accomplished this task in three weeks (based on an 8-hour workday, five days a week) while maintaining a similar or even better annotation quality. This achievement is attributed to three unique properties of our method: (1) label bias reduction using multiple pre-trained segmentation models, (2) effective error detection in the model predictions, and (3) attention guidance for annotators to make corrections on the most salient errors. Furthermore, we summarize the taxonomy of common errors made by AI algorithms and annotators. This allows for continuous refinement of both AI and annotations and significantly reduces the annotation costs required to create large-scale datasets for a wider variety of medical imaging tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge