Alan L. Yuille
Early and Prediagnostic Detection of Pancreatic Cancer from Computed Tomography
Jan 29, 2026Abstract:Pancreatic ductal adenocarcinoma (PDAC), one of the deadliest solid malignancies, is often detected at a late and inoperable stage. Retrospective reviews of prediagnostic CT scans, when conducted by expert radiologists aware that the patient later developed PDAC, frequently reveal lesions that were previously overlooked. To help detecting these lesions earlier, we developed an automated system named ePAI (early Pancreatic cancer detection with Artificial Intelligence). It was trained on data from 1,598 patients from a single medical center. In the internal test involving 1,009 patients, ePAI achieved an area under the receiver operating characteristic curve (AUC) of 0.939-0.999, a sensitivity of 95.3%, and a specificity of 98.7% for detecting small PDAC less than 2 cm in diameter, precisely localizing PDAC as small as 2 mm. In an external test involving 7,158 patients across 6 centers, ePAI achieved an AUC of 0.918-0.945, a sensitivity of 91.5%, and a specificity of 88.0%, precisely localizing PDAC as small as 5 mm. Importantly, ePAI detected PDACs on prediagnostic CT scans obtained 3 to 36 months before clinical diagnosis that had originally been overlooked by radiologists. It successfully detected and localized PDACs in 75 of 159 patients, with a median lead time of 347 days before clinical diagnosis. Our multi-reader study showed that ePAI significantly outperformed 30 board-certified radiologists by 50.3% (P < 0.05) in sensitivity while maintaining a comparable specificity of 95.4% in detecting PDACs early and prediagnostic. These findings suggest its potential of ePAI as an assistive tool to improve early detection of pancreatic cancer.
Auditing Significance, Metric Choice, and Demographic Fairness in Medical AI Challenges
Dec 22, 2025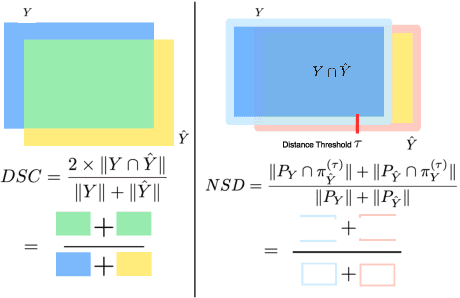
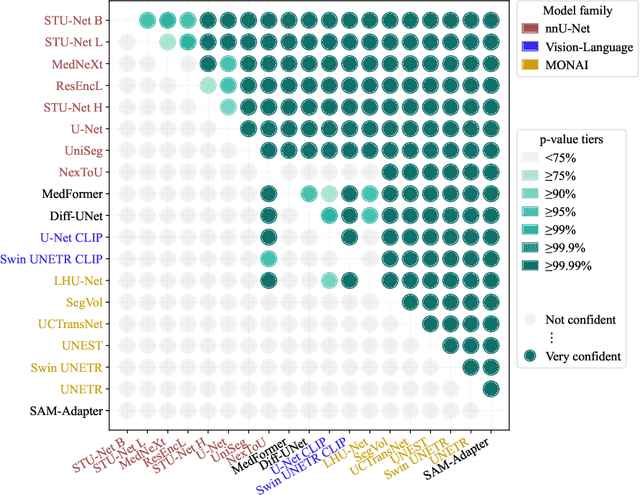
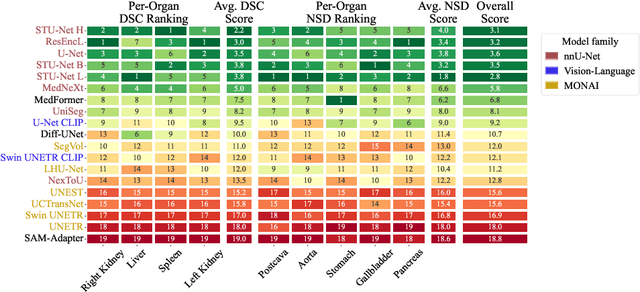
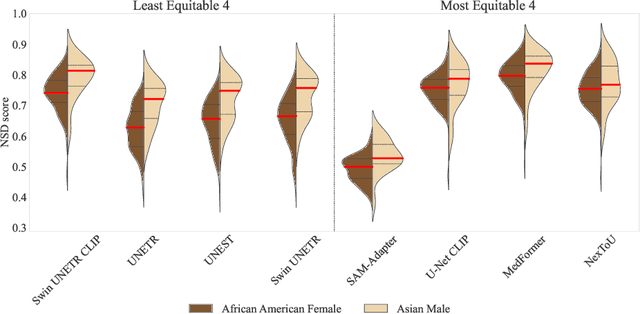
Abstract:Open challenges have become the de facto standard for comparative ranking of medical AI methods. Despite their importance, medical AI leaderboards exhibit three persistent limitations: (1) score gaps are rarely tested for statistical significance, so rank stability is unknown; (2) single averaged metrics are applied to every organ, hiding clinically important boundary errors; (3) performance across intersecting demographics is seldom reported, masking fairness and equity gaps. We introduce RankInsight, an open-source toolkit that seeks to address these limitations. RankInsight (1) computes pair-wise significance maps that show the nnU-Net family outperforms Vision-Language and MONAI submissions with high statistical certainty; (2) recomputes leaderboards with organ-appropriate metrics, reversing the order of the top four models when Dice is replaced by NSD for tubular structures; and (3) audits intersectional fairness, revealing that more than half of the MONAI-based entries have the largest gender-race discrepancy on our proprietary Johns Hopkins Hospital dataset. The RankInsight toolkit is publicly released and can be directly applied to past, ongoing, and future challenges. It enables organizers and participants to publish rankings that are statistically sound, clinically meaningful, and demographically fair.
See More, Change Less: Anatomy-Aware Diffusion for Contrast Enhancement
Dec 08, 2025Abstract:Image enhancement improves visual quality and helps reveal details that are hard to see in the original image. In medical imaging, it can support clinical decision-making, but current models often over-edit. This can distort organs, create false findings, and miss small tumors because these models do not understand anatomy or contrast dynamics. We propose SMILE, an anatomy-aware diffusion model that learns how organs are shaped and how they take up contrast. It enhances only clinically relevant regions while leaving all other areas unchanged. SMILE introduces three key ideas: (1) structure-aware supervision that follows true organ boundaries and contrast patterns; (2) registration-free learning that works directly with unaligned multi-phase CT scans; (3) unified inference that provides fast and consistent enhancement across all contrast phases. Across six external datasets, SMILE outperforms existing methods in image quality (14.2% higher SSIM, 20.6% higher PSNR, 50% better FID) and in clinical usefulness by producing anatomically accurate and diagnostically meaningful images. SMILE also improves cancer detection from non-contrast CT, raising the F1 score by up to 10 percent.
PanTS: The Pancreatic Tumor Segmentation Dataset
Jul 02, 2025Abstract:PanTS is a large-scale, multi-institutional dataset curated to advance research in pancreatic CT analysis. It contains 36,390 CT scans from 145 medical centers, with expert-validated, voxel-wise annotations of over 993,000 anatomical structures, covering pancreatic tumors, pancreas head, body, and tail, and 24 surrounding anatomical structures such as vascular/skeletal structures and abdominal/thoracic organs. Each scan includes metadata such as patient age, sex, diagnosis, contrast phase, in-plane spacing, slice thickness, etc. AI models trained on PanTS achieve significantly better performance in pancreatic tumor detection, localization, and segmentation compared to those trained on existing public datasets. Our analysis indicates that these gains are directly attributable to the 16x larger-scale tumor annotations and indirectly supported by the 24 additional surrounding anatomical structures. As the largest and most comprehensive resource of its kind, PanTS offers a new benchmark for developing and evaluating AI models in pancreatic CT analysis.
A Continual Learning-driven Model for Accurate and Generalizable Segmentation of Clinically Comprehensive and Fine-grained Whole-body Anatomies in CT
Mar 16, 2025Abstract:Precision medicine in the quantitative management of chronic diseases and oncology would be greatly improved if the Computed Tomography (CT) scan of any patient could be segmented, parsed and analyzed in a precise and detailed way. However, there is no such fully annotated CT dataset with all anatomies delineated for training because of the exceptionally high manual cost, the need for specialized clinical expertise, and the time required to finish the task. To this end, we proposed a novel continual learning-driven CT model that can segment complete anatomies presented using dozens of previously partially labeled datasets, dynamically expanding its capacity to segment new ones without compromising previously learned organ knowledge. Existing multi-dataset approaches are not able to dynamically segment new anatomies without catastrophic forgetting and would encounter optimization difficulty or infeasibility when segmenting hundreds of anatomies across the whole range of body regions. Our single unified CT segmentation model, CL-Net, can highly accurately segment a clinically comprehensive set of 235 fine-grained whole-body anatomies. Composed of a universal encoder, multiple optimized and pruned decoders, CL-Net is developed using 13,952 CT scans from 20 public and 16 private high-quality partially labeled CT datasets of various vendors, different contrast phases, and pathologies. Extensive evaluation demonstrates that CL-Net consistently outperforms the upper limit of an ensemble of 36 specialist nnUNets trained per dataset with the complexity of 5% model size and significantly surpasses the segmentation accuracy of recent leading Segment Anything-style medical image foundation models by large margins. Our continual learning-driven CL-Net model would lay a solid foundation to facilitate many downstream tasks of oncology and chronic diseases using the most widely adopted CT imaging.
ScaleMAI: Accelerating the Development of Trusted Datasets and AI Models
Jan 06, 2025

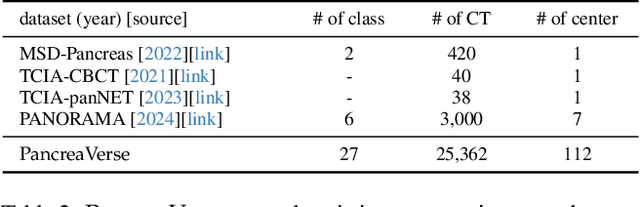

Abstract:Building trusted datasets is critical for transparent and responsible Medical AI (MAI) research, but creating even small, high-quality datasets can take years of effort from multidisciplinary teams. This process often delays AI benefits, as human-centric data creation and AI-centric model development are treated as separate, sequential steps. To overcome this, we propose ScaleMAI, an agent of AI-integrated data curation and annotation, allowing data quality and AI performance to improve in a self-reinforcing cycle and reducing development time from years to months. We adopt pancreatic tumor detection as an example. First, ScaleMAI progressively creates a dataset of 25,362 CT scans, including per-voxel annotations for benign/malignant tumors and 24 anatomical structures. Second, through progressive human-in-the-loop iterations, ScaleMAI provides Flagship AI Model that can approach the proficiency of expert annotators (30-year experience) in detecting pancreatic tumors. Flagship Model significantly outperforms models developed from smaller, fixed-quality datasets, with substantial gains in tumor detection (+14%), segmentation (+5%), and classification (72%) on three prestigious benchmarks. In summary, ScaleMAI transforms the speed, scale, and reliability of medical dataset creation, paving the way for a variety of impactful, data-driven applications.
Touchstone Benchmark: Are We on the Right Way for Evaluating AI Algorithms for Medical Segmentation?
Nov 06, 2024



Abstract:How can we test AI performance? This question seems trivial, but it isn't. Standard benchmarks often have problems such as in-distribution and small-size test sets, oversimplified metrics, unfair comparisons, and short-term outcome pressure. As a consequence, good performance on standard benchmarks does not guarantee success in real-world scenarios. To address these problems, we present Touchstone, a large-scale collaborative segmentation benchmark of 9 types of abdominal organs. This benchmark is based on 5,195 training CT scans from 76 hospitals around the world and 5,903 testing CT scans from 11 additional hospitals. This diverse test set enhances the statistical significance of benchmark results and rigorously evaluates AI algorithms across various out-of-distribution scenarios. We invited 14 inventors of 19 AI algorithms to train their algorithms, while our team, as a third party, independently evaluated these algorithms on three test sets. In addition, we also evaluated pre-existing AI frameworks--which, differing from algorithms, are more flexible and can support different algorithms--including MONAI from NVIDIA, nnU-Net from DKFZ, and numerous other open-source frameworks. We are committed to expanding this benchmark to encourage more innovation of AI algorithms for the medical domain.
Making Your First Choice: To Address Cold Start Problem in Vision Active Learning
Oct 05, 2022



Abstract:Active learning promises to improve annotation efficiency by iteratively selecting the most important data to be annotated first. However, we uncover a striking contradiction to this promise: active learning fails to select data as efficiently as random selection at the first few choices. We identify this as the cold start problem in vision active learning, caused by a biased and outlier initial query. This paper seeks to address the cold start problem by exploiting the three advantages of contrastive learning: (1) no annotation is required; (2) label diversity is ensured by pseudo-labels to mitigate bias; (3) typical data is determined by contrastive features to reduce outliers. Experiments are conducted on CIFAR-10-LT and three medical imaging datasets (i.e. Colon Pathology, Abdominal CT, and Blood Cell Microscope). Our initial query not only significantly outperforms existing active querying strategies but also surpasses random selection by a large margin. We foresee our solution to the cold start problem as a simple yet strong baseline to choose the initial query for vision active learning. Code is available: https://github.com/c-liangyu/CSVAL
External Attention Assisted Multi-Phase Splenic Vascular Injury Segmentation with Limited Data
Jan 04, 2022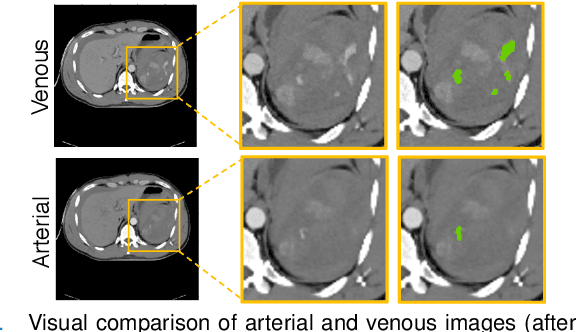
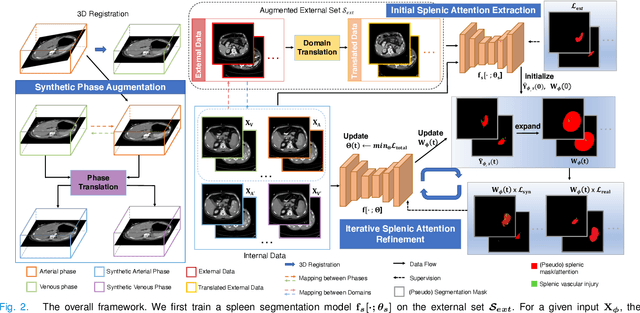
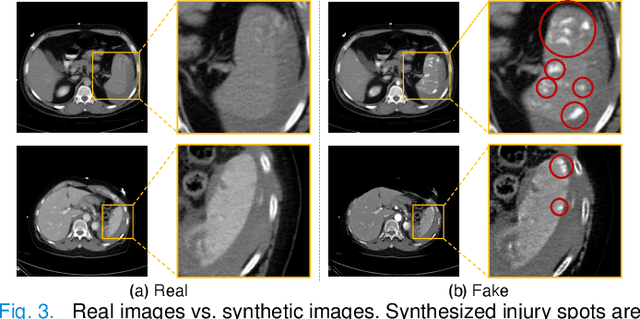
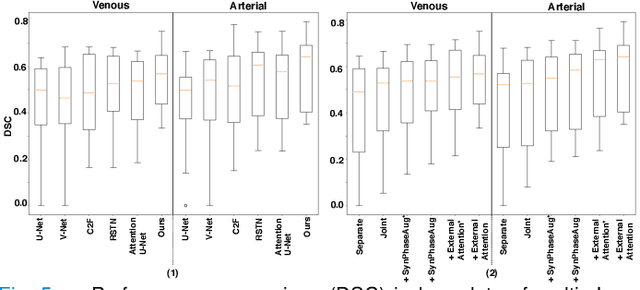
Abstract:The spleen is one of the most commonly injured solid organs in blunt abdominal trauma. The development of automatic segmentation systems from multi-phase CT for splenic vascular injury can augment severity grading for improving clinical decision support and outcome prediction. However, accurate segmentation of splenic vascular injury is challenging for the following reasons: 1) Splenic vascular injury can be highly variant in shape, texture, size, and overall appearance; and 2) Data acquisition is a complex and expensive procedure that requires intensive efforts from both data scientists and radiologists, which makes large-scale well-annotated datasets hard to acquire in general. In light of these challenges, we hereby design a novel framework for multi-phase splenic vascular injury segmentation, especially with limited data. On the one hand, we propose to leverage external data to mine pseudo splenic masks as the spatial attention, dubbed external attention, for guiding the segmentation of splenic vascular injury. On the other hand, we develop a synthetic phase augmentation module, which builds upon generative adversarial networks, for populating the internal data by fully leveraging the relation between different phases. By jointly enforcing external attention and populating internal data representation during training, our proposed method outperforms other competing methods and substantially improves the popular DeepLab-v3+ baseline by more than 7% in terms of average DSC, which confirms its effectiveness.
In-painting Radiography Images for Unsupervised Anomaly Detection
Nov 30, 2021



Abstract:We propose space-aware memory queues for in-painting and detecting anomalies from radiography images (abbreviated as SQUID). Radiography imaging protocols focus on particular body regions, therefore producing images of great similarity and yielding recurrent anatomical structures across patients. To exploit this structured information, our SQUID consists of a new Memory Queue and a novel in-painting block in the feature space. We show that SQUID can taxonomize the ingrained anatomical structures into recurrent patterns; and in the inference, SQUID can identify anomalies (unseen/modified patterns) in the image. SQUID surpasses the state of the art in unsupervised anomaly detection by over 5 points on two chest X-ray benchmark datasets. Additionally, we have created a new dataset (DigitAnatomy), which synthesizes the spatial correlation and consistent shape in chest anatomy. We hope DigitAnatomy can prompt the development, evaluation, and interpretability of anomaly detection methods, particularly for radiography imaging.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge