Zi Li
OncoReg: Medical Image Registration for Oncological Challenges
Apr 01, 2025Abstract:In modern cancer research, the vast volume of medical data generated is often underutilised due to challenges related to patient privacy. The OncoReg Challenge addresses this issue by enabling researchers to develop and validate image registration methods through a two-phase framework that ensures patient privacy while fostering the development of more generalisable AI models. Phase one involves working with a publicly available dataset, while phase two focuses on training models on a private dataset within secure hospital networks. OncoReg builds upon the foundation established by the Learn2Reg Challenge by incorporating the registration of interventional cone-beam computed tomography (CBCT) with standard planning fan-beam CT (FBCT) images in radiotherapy. Accurate image registration is crucial in oncology, particularly for dynamic treatment adjustments in image-guided radiotherapy, where precise alignment is necessary to minimise radiation exposure to healthy tissues while effectively targeting tumours. This work details the methodology and data behind the OncoReg Challenge and provides a comprehensive analysis of the competition entries and results. Findings reveal that feature extraction plays a pivotal role in this registration task. A new method emerging from this challenge demonstrated its versatility, while established approaches continue to perform comparably to newer techniques. Both deep learning and classical approaches still play significant roles in image registration, with the combination of methods - particularly in feature extraction - proving most effective.
UniReg: Foundation Model for Controllable Medical Image Registration
Mar 17, 2025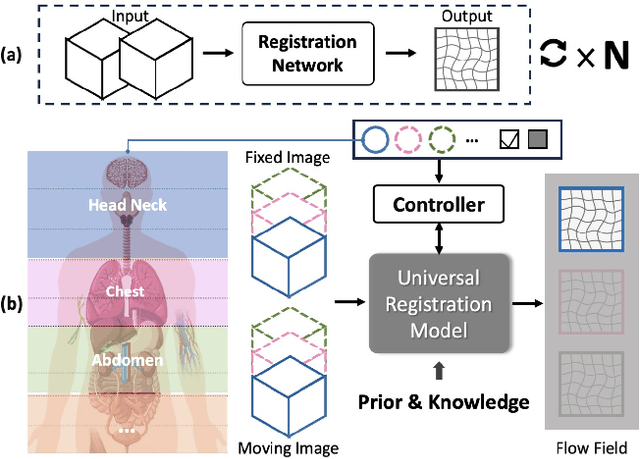
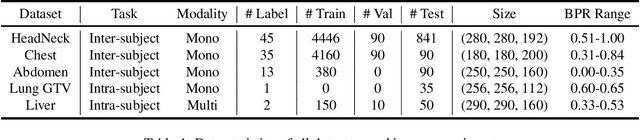
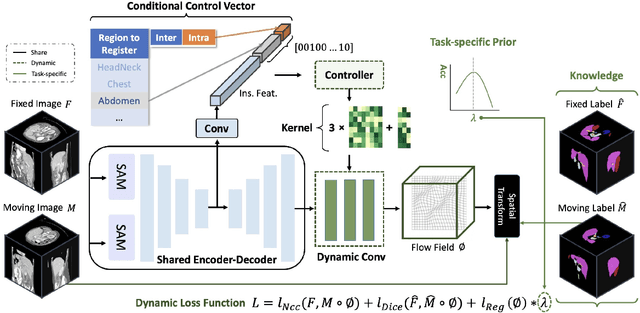
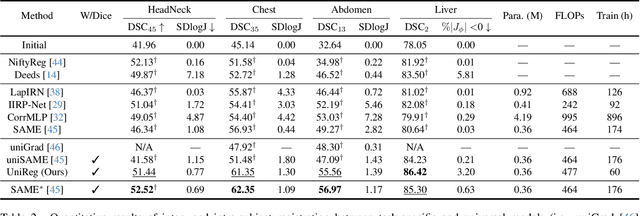
Abstract:Learning-based medical image registration has achieved performance parity with conventional methods while demonstrating a substantial advantage in computational efficiency. However, learning-based registration approaches lack generalizability across diverse clinical scenarios, requiring the laborious development of multiple isolated networks for specific registration tasks, e.g., inter-/intra-subject registration or organ-specific alignment. % To overcome this limitation, we propose \textbf{UniReg}, the first interactive foundation model for medical image registration, which combines the precision advantages of task-specific learning methods with the generalization of traditional optimization methods. Our key innovation is a unified framework for diverse registration scenarios, achieved through a conditional deformation field estimation within a unified registration model. This is realized through a dynamic learning paradigm that explicitly encodes: (1) anatomical structure priors, (2) registration type constraints (inter/intra-subject), and (3) instance-specific features, enabling the generation of scenario-optimal deformation fields. % Through comprehensive experiments encompassing $90$ anatomical structures at different body regions, our UniReg model demonstrates comparable performance with contemporary state-of-the-art methodologies while achieving ~50\% reduction in required training iterations relative to the conventional learning-based paradigm. This optimization contributes to a significant reduction in computational resources, such as training time. Code and model will be available.
A Continual Learning-driven Model for Accurate and Generalizable Segmentation of Clinically Comprehensive and Fine-grained Whole-body Anatomies in CT
Mar 16, 2025Abstract:Precision medicine in the quantitative management of chronic diseases and oncology would be greatly improved if the Computed Tomography (CT) scan of any patient could be segmented, parsed and analyzed in a precise and detailed way. However, there is no such fully annotated CT dataset with all anatomies delineated for training because of the exceptionally high manual cost, the need for specialized clinical expertise, and the time required to finish the task. To this end, we proposed a novel continual learning-driven CT model that can segment complete anatomies presented using dozens of previously partially labeled datasets, dynamically expanding its capacity to segment new ones without compromising previously learned organ knowledge. Existing multi-dataset approaches are not able to dynamically segment new anatomies without catastrophic forgetting and would encounter optimization difficulty or infeasibility when segmenting hundreds of anatomies across the whole range of body regions. Our single unified CT segmentation model, CL-Net, can highly accurately segment a clinically comprehensive set of 235 fine-grained whole-body anatomies. Composed of a universal encoder, multiple optimized and pruned decoders, CL-Net is developed using 13,952 CT scans from 20 public and 16 private high-quality partially labeled CT datasets of various vendors, different contrast phases, and pathologies. Extensive evaluation demonstrates that CL-Net consistently outperforms the upper limit of an ensemble of 36 specialist nnUNets trained per dataset with the complexity of 5% model size and significantly surpasses the segmentation accuracy of recent leading Segment Anything-style medical image foundation models by large margins. Our continual learning-driven CL-Net model would lay a solid foundation to facilitate many downstream tasks of oncology and chronic diseases using the most widely adopted CT imaging.
LicenseGPT: A Fine-tuned Foundation Model for Publicly Available Dataset License Compliance
Dec 30, 2024
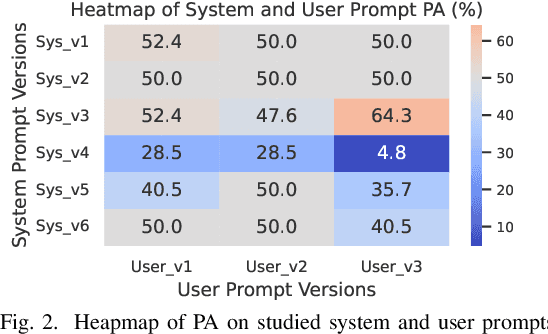

Abstract:Dataset license compliance is a critical yet complex aspect of developing commercial AI products, particularly with the increasing use of publicly available datasets. Ambiguities in dataset licenses pose significant legal risks, making it challenging even for software IP lawyers to accurately interpret rights and obligations. In this paper, we introduce LicenseGPT, a fine-tuned foundation model (FM) specifically designed for dataset license compliance analysis. We first evaluate existing legal FMs (i.e., FMs specialized in understanding and processing legal texts) and find that the best-performing model achieves a Prediction Agreement (PA) of only 43.75%. LicenseGPT, fine-tuned on a curated dataset of 500 licenses annotated by legal experts, significantly improves PA to 64.30%, outperforming both legal and general-purpose FMs. Through an A/B test and user study with software IP lawyers, we demonstrate that LicenseGPT reduces analysis time by 94.44%, from 108 seconds to 6 seconds per license, without compromising accuracy. Software IP lawyers perceive LicenseGPT as a valuable supplementary tool that enhances efficiency while acknowledging the need for human oversight in complex cases. Our work underscores the potential of specialized AI tools in legal practice and offers a publicly available resource for practitioners and researchers.
Leveraging Semantic Asymmetry for Precise Gross Tumor Volume Segmentation of Nasopharyngeal Carcinoma in Planning CT
Nov 27, 2024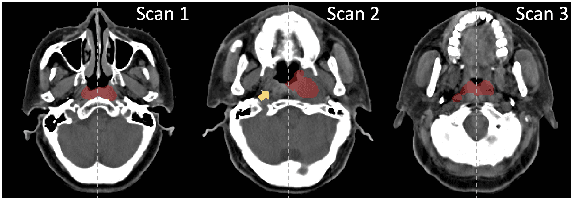
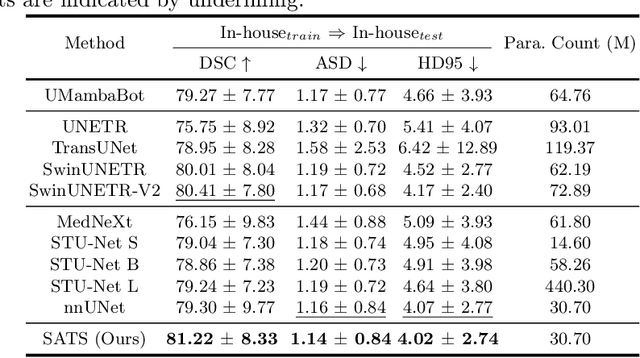
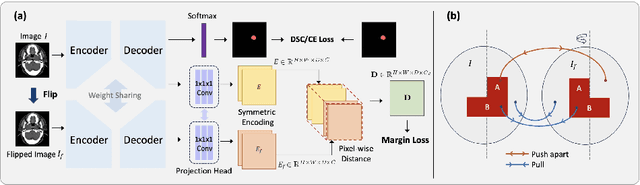
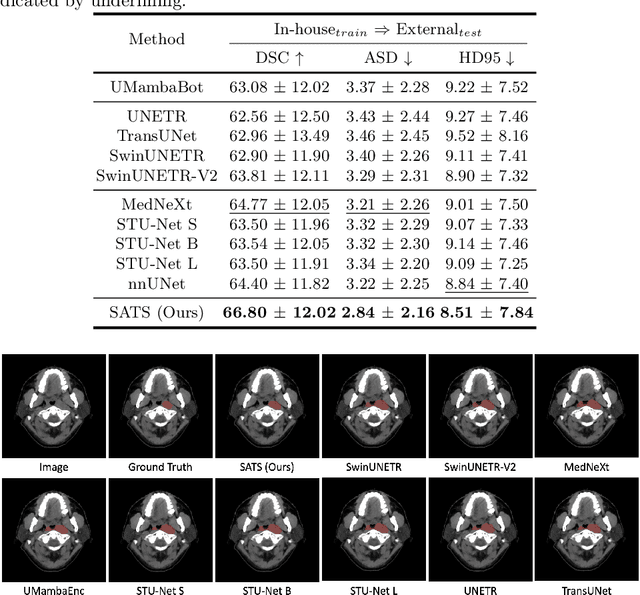
Abstract:In the radiation therapy of nasopharyngeal carcinoma (NPC), clinicians typically delineate the gross tumor volume (GTV) using non-contrast planning computed tomography to ensure accurate radiation dose delivery. However, the low contrast between tumors and adjacent normal tissues necessitates that radiation oncologists manually delineate the tumors, often relying on diagnostic MRI for guidance. % In this study, we propose a novel approach to directly segment NPC gross tumors on non-contrast planning CT images, circumventing potential registration errors when aligning MRI or MRI-derived tumor masks to planning CT. To address the low contrast issues between tumors and adjacent normal structures in planning CT, we introduce a 3D Semantic Asymmetry Tumor segmentation (SATs) method. Specifically, we posit that a healthy nasopharyngeal region is characteristically bilaterally symmetric, whereas the emergence of nasopharyngeal carcinoma disrupts this symmetry. Then, we propose a Siamese contrastive learning segmentation framework that minimizes the voxel-wise distance between original and flipped areas without tumor and encourages a larger distance between original and flipped areas with tumor. Thus, our approach enhances the sensitivity of features to semantic asymmetries. % Extensive experiments demonstrate that the proposed SATs achieves the leading NPC GTV segmentation performance in both internal and external testing, \emph{e.g.}, with at least 2\% absolute Dice score improvement and 12\% average distance error reduction when compared to other state-of-the-art methods in the external testing.
Rapid and Accurate Diagnosis of Acute Aortic Syndrome using Non-contrast CT: A Large-scale, Retrospective, Multi-center and AI-based Study
Jun 25, 2024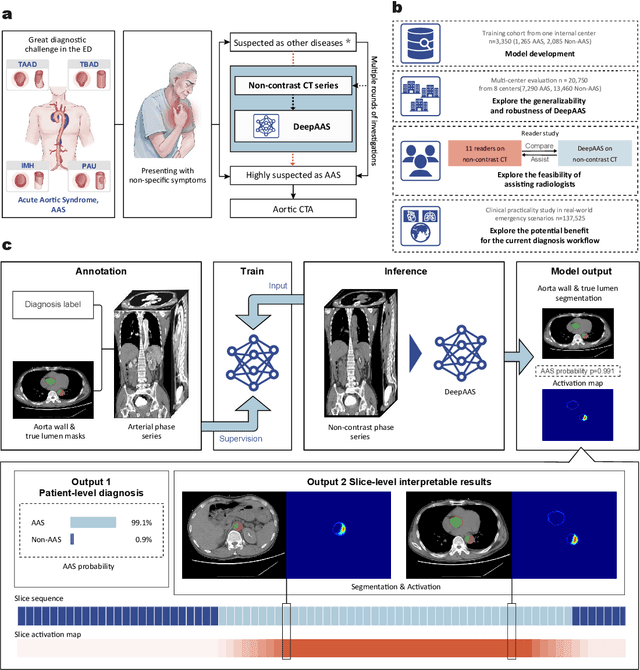
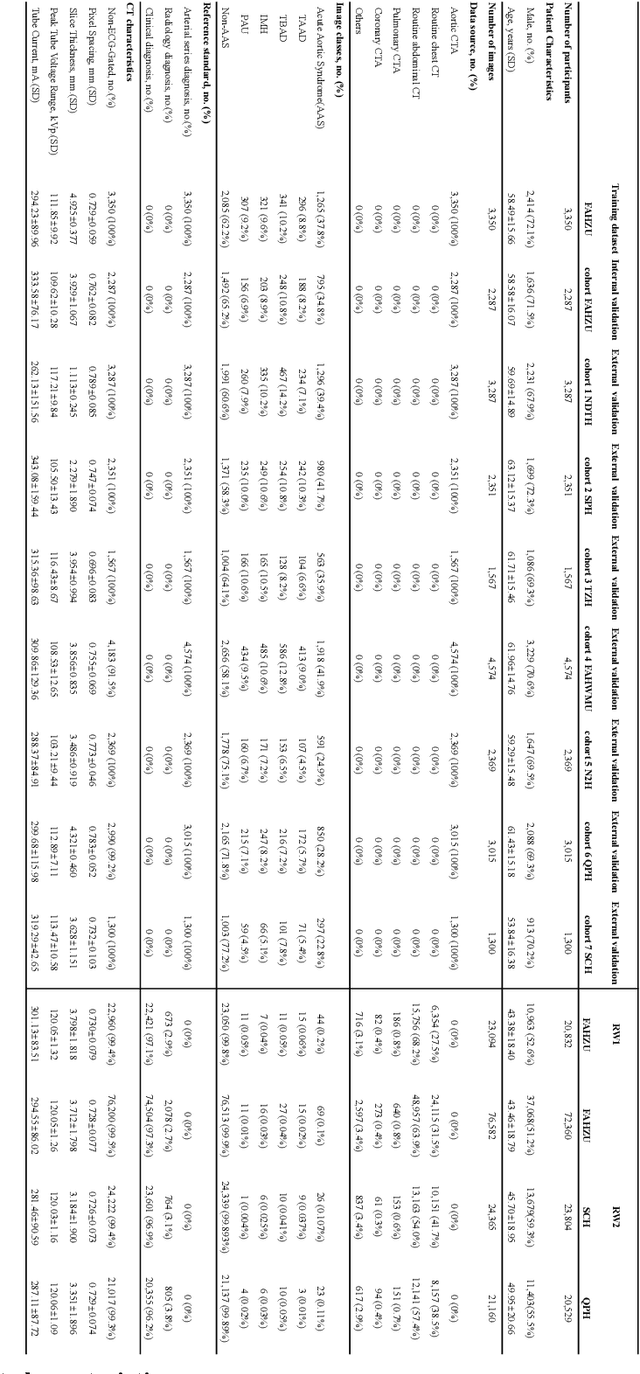
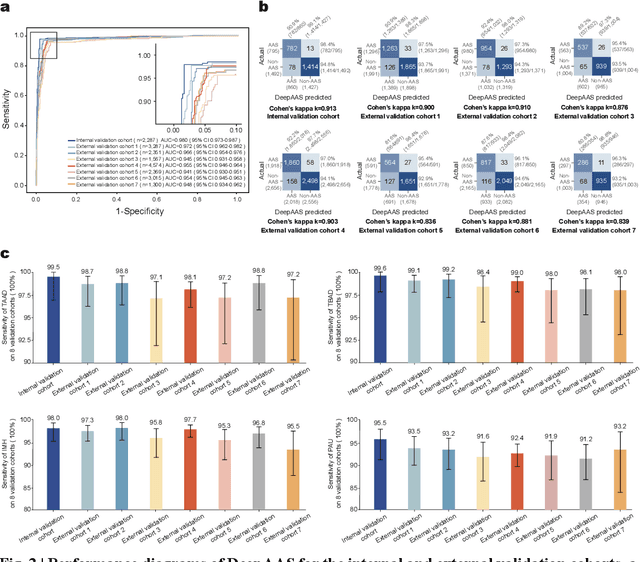
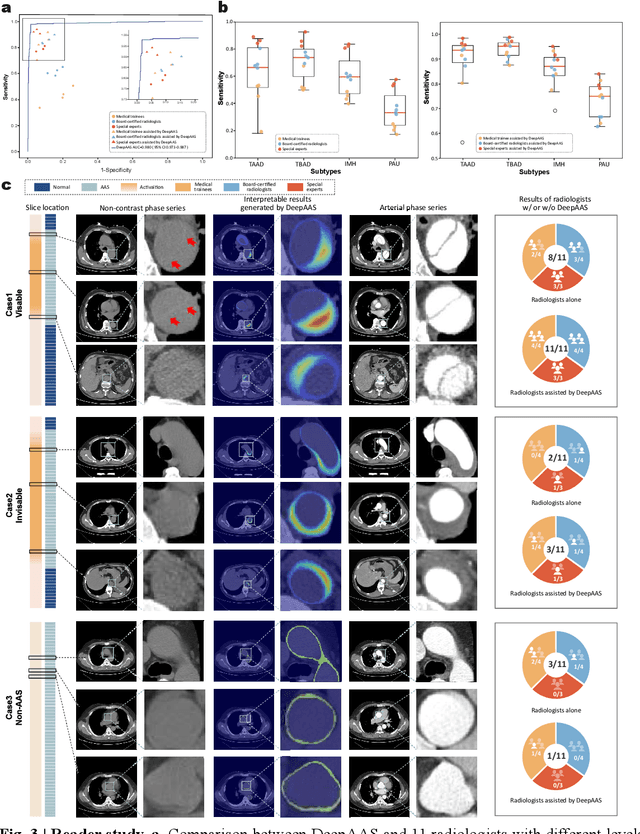
Abstract:Chest pain symptoms are highly prevalent in emergency departments (EDs), where acute aortic syndrome (AAS) is a catastrophic cardiovascular emergency with a high fatality rate, especially when timely and accurate treatment is not administered. However, current triage practices in the ED can cause up to approximately half of patients with AAS to have an initially missed diagnosis or be misdiagnosed as having other acute chest pain conditions. Subsequently, these AAS patients will undergo clinically inaccurate or suboptimal differential diagnosis. Fortunately, even under these suboptimal protocols, nearly all these patients underwent non-contrast CT covering the aorta anatomy at the early stage of differential diagnosis. In this study, we developed an artificial intelligence model (DeepAAS) using non-contrast CT, which is highly accurate for identifying AAS and provides interpretable results to assist in clinical decision-making. Performance was assessed in two major phases: a multi-center retrospective study (n = 20,750) and an exploration in real-world emergency scenarios (n = 137,525). In the multi-center cohort, DeepAAS achieved a mean area under the receiver operating characteristic curve of 0.958 (95% CI 0.950-0.967). In the real-world cohort, DeepAAS detected 109 AAS patients with misguided initial suspicion, achieving 92.6% (95% CI 76.2%-97.5%) in mean sensitivity and 99.2% (95% CI 99.1%-99.3%) in mean specificity. Our AI model performed well on non-contrast CT at all applicable early stages of differential diagnosis workflows, effectively reduced the overall missed diagnosis and misdiagnosis rate from 48.8% to 4.8% and shortened the diagnosis time for patients with misguided initial suspicion from an average of 681.8 (74-11,820) mins to 68.5 (23-195) mins. DeepAAS could effectively fill the gap in the current clinical workflow without requiring additional tests.
Bootstrapping Chest CT Image Understanding by Distilling Knowledge from X-ray Expert Models
Apr 07, 2024



Abstract:Radiologists highly desire fully automated versatile AI for medical imaging interpretation. However, the lack of extensively annotated large-scale multi-disease datasets has hindered the achievement of this goal. In this paper, we explore the feasibility of leveraging language as a naturally high-quality supervision for chest CT imaging. In light of the limited availability of image-report pairs, we bootstrap the understanding of 3D chest CT images by distilling chest-related diagnostic knowledge from an extensively pre-trained 2D X-ray expert model. Specifically, we propose a language-guided retrieval method to match each 3D CT image with its semantically closest 2D X-ray image, and perform pair-wise and semantic relation knowledge distillation. Subsequently, we use contrastive learning to align images and reports within the same patient while distinguishing them from the other patients. However, the challenge arises when patients have similar semantic diagnoses, such as healthy patients, potentially confusing if treated as negatives. We introduce a robust contrastive learning that identifies and corrects these false negatives. We train our model with over 12,000 pairs of chest CT images and radiology reports. Extensive experiments across multiple scenarios, including zero-shot learning, report generation, and fine-tuning processes, demonstrate the model's feasibility in interpreting chest CT images.
Modality-Agnostic Structural Image Representation Learning for Deformable Multi-Modality Medical Image Registration
Feb 29, 2024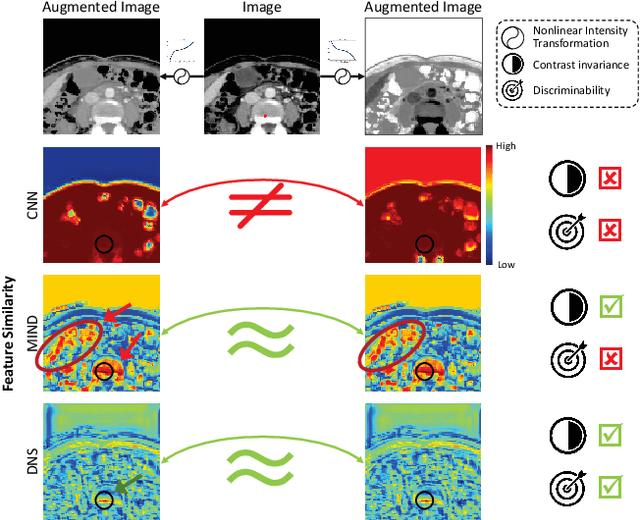
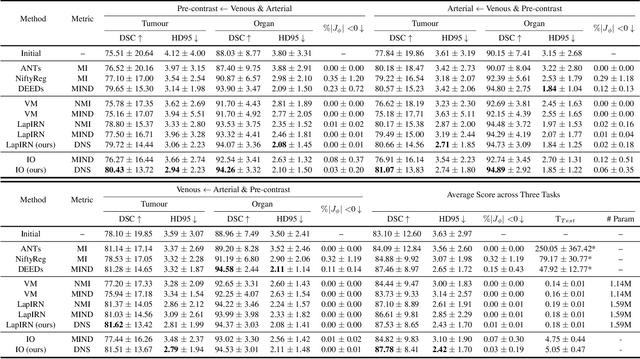
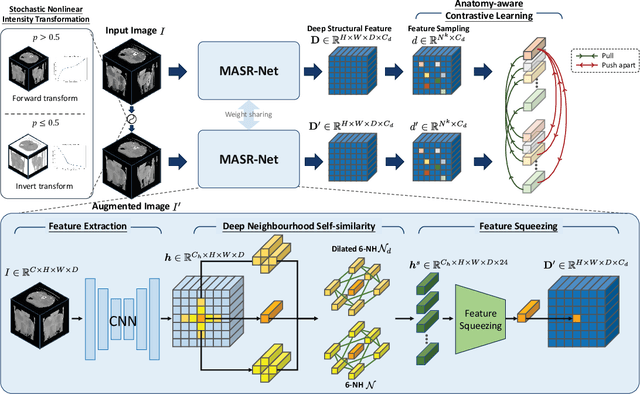
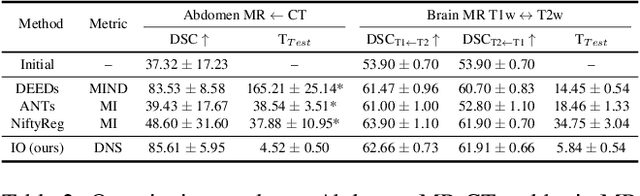
Abstract:Establishing dense anatomical correspondence across distinct imaging modalities is a foundational yet challenging procedure for numerous medical image analysis studies and image-guided radiotherapy. Existing multi-modality image registration algorithms rely on statistical-based similarity measures or local structural image representations. However, the former is sensitive to locally varying noise, while the latter is not discriminative enough to cope with complex anatomical structures in multimodal scans, causing ambiguity in determining the anatomical correspondence across scans with different modalities. In this paper, we propose a modality-agnostic structural representation learning method, which leverages Deep Neighbourhood Self-similarity (DNS) and anatomy-aware contrastive learning to learn discriminative and contrast-invariance deep structural image representations (DSIR) without the need for anatomical delineations or pre-aligned training images. We evaluate our method on multiphase CT, abdomen MR-CT, and brain MR T1w-T2w registration. Comprehensive results demonstrate that our method is superior to the conventional local structural representation and statistical-based similarity measures in terms of discriminability and accuracy.
TAROT: A Hierarchical Framework with Multitask Co-Pretraining on Semi-Structured Data towards Effective Person-Job Fit
Jan 17, 2024Abstract:Person-job fit is an essential part of online recruitment platforms in serving various downstream applications like Job Search and Candidate Recommendation. Recently, pretrained large language models have further enhanced the effectiveness by leveraging richer textual information in user profiles and job descriptions apart from user behavior features and job metadata. However, the general domain-oriented design struggles to capture the unique structural information within user profiles and job descriptions, leading to a loss of latent semantic correlations. We propose TAROT, a hierarchical multitask co-pretraining framework, to better utilize structural and semantic information for informative text embeddings. TAROT targets semi-structured text in profiles and jobs, and it is co-pretained with multi-grained pretraining tasks to constrain the acquired semantic information at each level. Experiments on a real-world LinkedIn dataset show significant performance improvements, proving its effectiveness in person-job fit tasks.
Professional Network Matters: Connections Empower Person-Job Fit
Dec 19, 2023Abstract:Online recruitment platforms typically employ Person-Job Fit models in the core service that automatically match suitable job seekers with appropriate job positions. While existing works leverage historical or contextual information, they often disregard a crucial aspect: job seekers' social relationships in professional networks. This paper emphasizes the importance of incorporating professional networks into the Person-Job Fit model. Our innovative approach consists of two stages: (1) defining a Workplace Heterogeneous Information Network (WHIN) to capture heterogeneous knowledge, including professional connections and pre-training representations of various entities using a heterogeneous graph neural network; (2) designing a Contextual Social Attention Graph Neural Network (CSAGNN) that supplements users' missing information with professional connections' contextual information. We introduce a job-specific attention mechanism in CSAGNN to handle noisy professional networks, leveraging pre-trained entity representations from WHIN. We demonstrate the effectiveness of our approach through experimental evaluations conducted across three real-world recruitment datasets from LinkedIn, showing superior performance compared to baseline models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge