Ke Yan
Non-Contrast CT Esophageal Varices Grading through Clinical Prior-Enhanced Multi-Organ Analysis
Dec 22, 2025Abstract:Esophageal varices (EV) represent a critical complication of portal hypertension, affecting approximately 60% of cirrhosis patients with a significant bleeding risk of ~30%. While traditionally diagnosed through invasive endoscopy, non-contrast computed tomography (NCCT) presents a potential non-invasive alternative that has yet to be fully utilized in clinical practice. We present Multi-Organ-COhesion Network++ (MOON++), a novel multimodal framework that enhances EV assessment through comprehensive analysis of NCCT scans. Inspired by clinical evidence correlating organ volumetric relationships with liver disease severity, MOON++ synthesizes imaging characteristics of the esophagus, liver, and spleen through multimodal learning. We evaluated our approach using 1,631 patients, those with endoscopically confirmed EV were classified into four severity grades. Validation in 239 patient cases and independent testing in 289 cases demonstrate superior performance compared to conventional single organ methods, achieving an AUC of 0.894 versus 0.803 for the severe grade EV classification (G3 versus <G3) and 0.921 versus 0.793 for the differentiation of moderate to severe grades (>=G2 versus <G2). We conducted a reader study involving experienced radiologists to further validate the performance of MOON++. To our knowledge, MOON++ represents the first comprehensive multi-organ NCCT analysis framework incorporating clinical knowledge priors for EV assessment, potentially offering a promising non-invasive diagnostic alternative.
D2Pruner: Debiased Importance and Structural Diversity for MLLM Token Pruning
Dec 22, 2025



Abstract:Processing long visual token sequences poses a significant computational burden on Multimodal Large Language Models (MLLMs). While token pruning offers a path to acceleration, we find that current methods, while adequate for general understanding, catastrophically fail on fine-grained localization tasks. We attribute this failure to the inherent flaws of the two prevailing strategies: importance-based methods suffer from a strong positional bias, an inherent model artifact that distracts from semantic content, while diversity-based methods exhibit structural blindness, disregarding the user's prompt and spatial redundancy. To address this, we introduce D2Pruner, a framework that rectifies these issues by uniquely combining debiased importance with a structural pruning mechanism. Our method first secures a core set of the most critical tokens as pivots based on a debiased attention score. It then performs a Maximal Independent Set (MIS) selection on the remaining tokens, which are modeled on a hybrid graph where edges signify spatial proximity and semantic similarity. This process iteratively preserves the most important and available token while removing its neighbors, ensuring that the supplementary tokens are chosen to maximize importance and diversity. Extensive experiments demonstrate that D2Pruner has exceptional efficiency and fidelity. Applied to LLaVA-1.5-7B for general understanding tasks, it reduces FLOPs by 74.2\% while retaining 99.2\% of its original performance. Furthermore, in challenging localization benchmarks with InternVL-2.5-8B, it maintains 85.7\% performance at a 90\% token reduction rate, marking a significant advancement with up to 63. 53\% improvement over existing methods.
Extracting Events Like Code: A Multi-Agent Programming Framework for Zero-Shot Event Extraction
Nov 17, 2025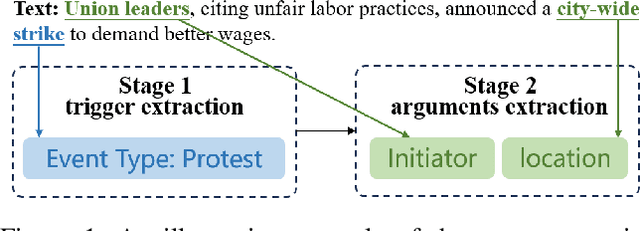
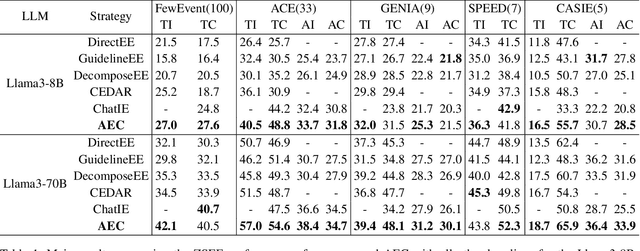
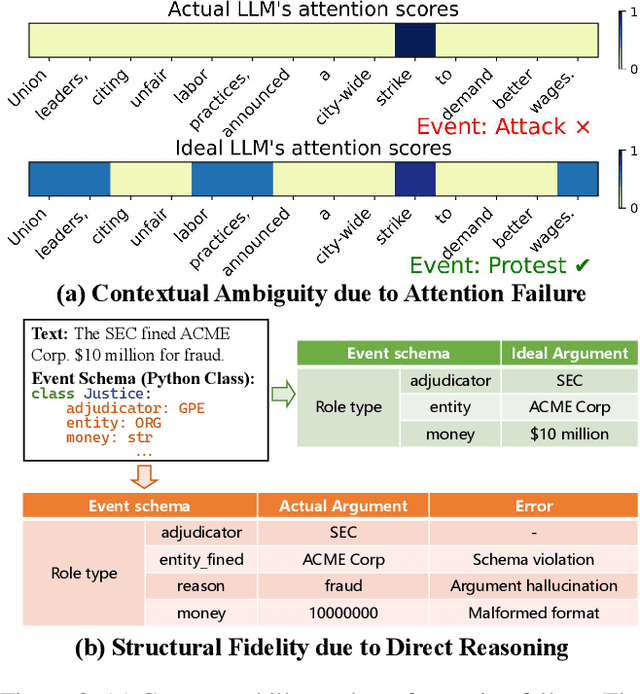
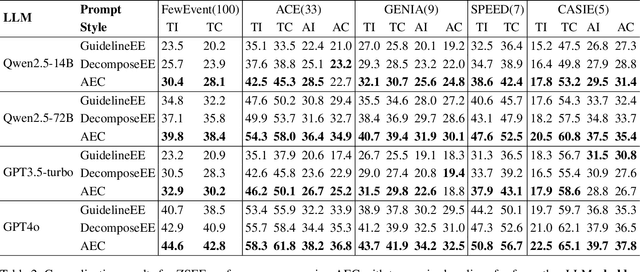
Abstract:Zero-shot event extraction (ZSEE) remains a significant challenge for large language models (LLMs) due to the need for complex reasoning and domain-specific understanding. Direct prompting often yields incomplete or structurally invalid outputs--such as misclassified triggers, missing arguments, and schema violations. To address these limitations, we present Agent-Event-Coder (AEC), a novel multi-agent framework that treats event extraction like software engineering: as a structured, iterative code-generation process. AEC decomposes ZSEE into specialized subtasks--retrieval, planning, coding, and verification--each handled by a dedicated LLM agent. Event schemas are represented as executable class definitions, enabling deterministic validation and precise feedback via a verification agent. This programming-inspired approach allows for systematic disambiguation and schema enforcement through iterative refinement. By leveraging collaborative agent workflows, AEC enables LLMs to produce precise, complete, and schema-consistent extractions in zero-shot settings. Experiments across five diverse domains and six LLMs demonstrate that AEC consistently outperforms prior zero-shot baselines, showcasing the power of treating event extraction like code generation. The code and data are released on https://github.com/UESTC-GQJ/Agent-Event-Coder.
ITPP: Learning Disentangled Event Dynamics in Marked Temporal Point Processes
Nov 08, 2025Abstract:Marked Temporal Point Processes (MTPPs) provide a principled framework for modeling asynchronous event sequences by conditioning on the history of past events. However, most existing MTPP models rely on channel-mixing strategies that encode information from different event types into a single, fixed-size latent representation. This entanglement can obscure type-specific dynamics, leading to performance degradation and increased risk of overfitting. In this work, we introduce ITPP, a novel channel-independent architecture for MTPP modeling that decouples event type information using an encoder-decoder framework with an ODE-based backbone. Central to ITPP is a type-aware inverted self-attention mechanism, designed to explicitly model inter-channel correlations among heterogeneous event types. This architecture enhances effectiveness and robustness while reducing overfitting. Comprehensive experiments on multiple real-world and synthetic datasets demonstrate that ITPP consistently outperforms state-of-the-art MTPP models in both predictive accuracy and generalization.
MUSE: Multi-Scale Dense Self-Distillation for Nucleus Detection and Classification
Nov 07, 2025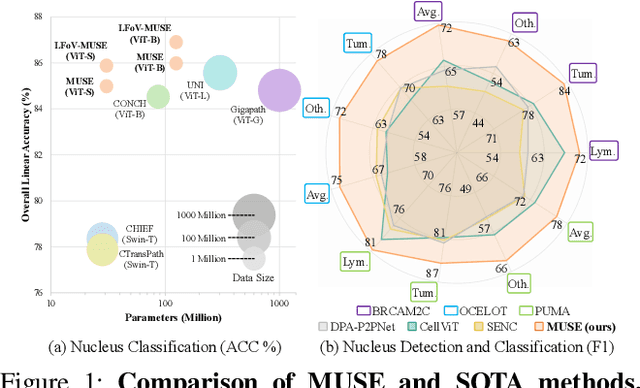
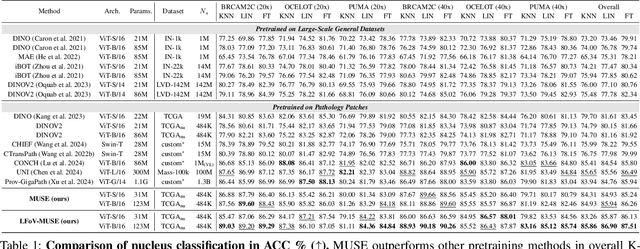
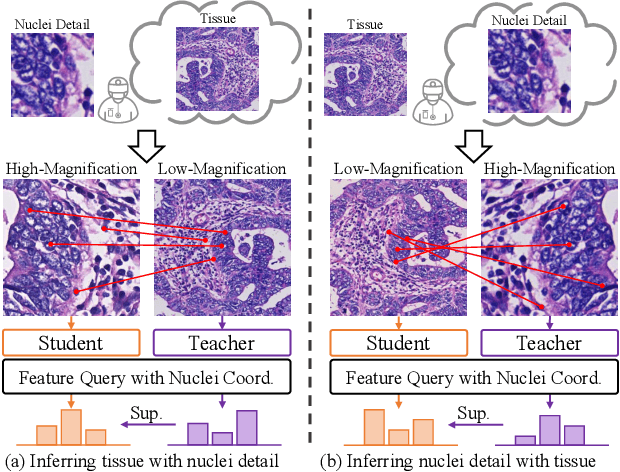
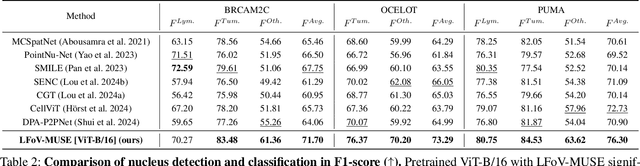
Abstract:Nucleus detection and classification (NDC) in histopathology analysis is a fundamental task that underpins a wide range of high-level pathology applications. However, existing methods heavily rely on labor-intensive nucleus-level annotations and struggle to fully exploit large-scale unlabeled data for learning discriminative nucleus representations. In this work, we propose MUSE (MUlti-scale denSE self-distillation), a novel self-supervised learning method tailored for NDC. At its core is NuLo (Nucleus-based Local self-distillation), a coordinate-guided mechanism that enables flexible local self-distillation based on predicted nucleus positions. By removing the need for strict spatial alignment between augmented views, NuLo allows critical cross-scale alignment, thus unlocking the capacity of models for fine-grained nucleus-level representation. To support MUSE, we design a simple yet effective encoder-decoder architecture and a large field-of-view semi-supervised fine-tuning strategy that together maximize the value of unlabeled pathology images. Extensive experiments on three widely used benchmarks demonstrate that MUSE effectively addresses the core challenges of histopathological NDC. The resulting models not only surpass state-of-the-art supervised baselines but also outperform generic pathology foundation models.
Towards Rationale-Answer Alignment of LVLMs via Self-Rationale Calibration
Sep 17, 2025Abstract:Large Vision-Language Models (LVLMs) have manifested strong visual question answering capability. However, they still struggle with aligning the rationale and the generated answer, leading to inconsistent reasoning and incorrect responses. To this end, this paper introduces the Self-Rationale Calibration (SRC) framework to iteratively calibrate the alignment between rationales and answers. SRC begins by employing a lightweight "rationale fine-tuning" approach, which modifies the model's response format to require a rationale before deriving an answer without explicit prompts. Next, SRC searches for a diverse set of candidate responses from the fine-tuned LVLMs for each sample, followed by a proposed pairwise scoring strategy using a tailored scoring model, R-Scorer, to evaluate both rationale quality and factual consistency of candidates. Based on a confidence-weighted preference curation process, SRC decouples the alignment calibration into a preference fine-tuning manner, leading to significant improvements of LVLMs in perception, reasoning, and generalization across multiple benchmarks. Our results emphasize the rationale-oriented alignment in exploring the potential of LVLMs.
VISA: Group-wise Visual Token Selection and Aggregation via Graph Summarization for Efficient MLLMs Inference
Aug 25, 2025Abstract:In this study, we introduce a novel method called group-wise \textbf{VI}sual token \textbf{S}election and \textbf{A}ggregation (VISA) to address the issue of inefficient inference stemming from excessive visual tokens in multimoal large language models (MLLMs). Compared with previous token pruning approaches, our method can preserve more visual information while compressing visual tokens. We first propose a graph-based visual token aggregation (VTA) module. VTA treats each visual token as a node, forming a graph based on semantic similarity among visual tokens. It then aggregates information from removed tokens into kept tokens based on this graph, producing a more compact visual token representation. Additionally, we introduce a group-wise token selection strategy (GTS) to divide visual tokens into kept and removed ones, guided by text tokens from the final layers of each group. This strategy progressively aggregates visual information, enhancing the stability of the visual information extraction process. We conduct comprehensive experiments on LLaVA-1.5, LLaVA-NeXT, and Video-LLaVA across various benchmarks to validate the efficacy of VISA. Our method consistently outperforms previous methods, achieving a superior trade-off between model performance and inference speed. The code is available at https://github.com/mobiushy/VISA.
AIGI-Holmes: Towards Explainable and Generalizable AI-Generated Image Detection via Multimodal Large Language Models
Jul 03, 2025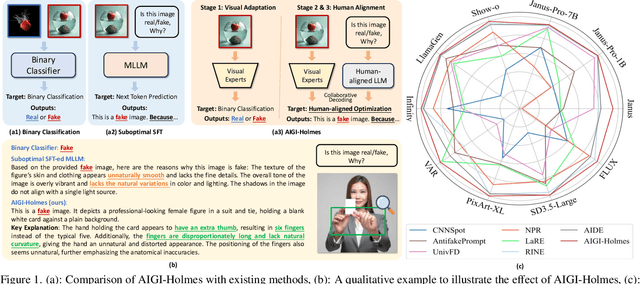
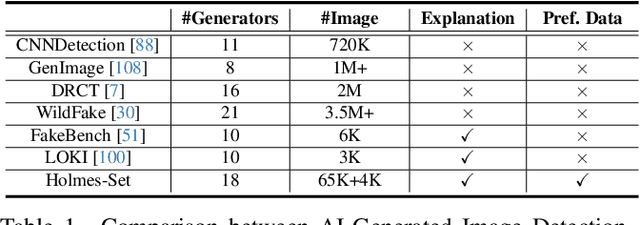
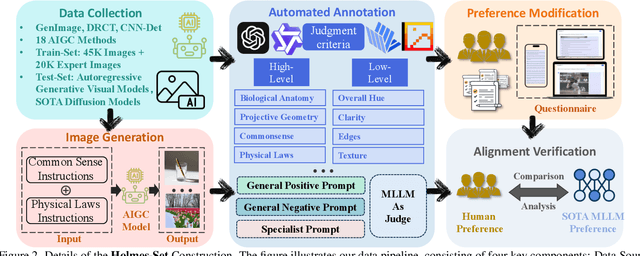
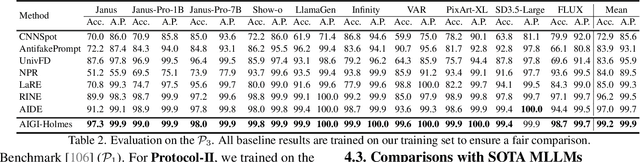
Abstract:The rapid development of AI-generated content (AIGC) technology has led to the misuse of highly realistic AI-generated images (AIGI) in spreading misinformation, posing a threat to public information security. Although existing AIGI detection techniques are generally effective, they face two issues: 1) a lack of human-verifiable explanations, and 2) a lack of generalization in the latest generation technology. To address these issues, we introduce a large-scale and comprehensive dataset, Holmes-Set, which includes the Holmes-SFTSet, an instruction-tuning dataset with explanations on whether images are AI-generated, and the Holmes-DPOSet, a human-aligned preference dataset. Our work introduces an efficient data annotation method called the Multi-Expert Jury, enhancing data generation through structured MLLM explanations and quality control via cross-model evaluation, expert defect filtering, and human preference modification. In addition, we propose Holmes Pipeline, a meticulously designed three-stage training framework comprising visual expert pre-training, supervised fine-tuning, and direct preference optimization. Holmes Pipeline adapts multimodal large language models (MLLMs) for AIGI detection while generating human-verifiable and human-aligned explanations, ultimately yielding our model AIGI-Holmes. During the inference stage, we introduce a collaborative decoding strategy that integrates the model perception of the visual expert with the semantic reasoning of MLLMs, further enhancing the generalization capabilities. Extensive experiments on three benchmarks validate the effectiveness of our AIGI-Holmes.
Antidote: A Unified Framework for Mitigating LVLM Hallucinations in Counterfactual Presupposition and Object Perception
Apr 29, 2025



Abstract:Large Vision-Language Models (LVLMs) have achieved impressive results across various cross-modal tasks. However, hallucinations, i.e., the models generating counterfactual responses, remain a challenge. Though recent studies have attempted to alleviate object perception hallucinations, they focus on the models' response generation, and overlooking the task question itself. This paper discusses the vulnerability of LVLMs in solving counterfactual presupposition questions (CPQs), where the models are prone to accept the presuppositions of counterfactual objects and produce severe hallucinatory responses. To this end, we introduce "Antidote", a unified, synthetic data-driven post-training framework for mitigating both types of hallucination above. It leverages synthetic data to incorporate factual priors into questions to achieve self-correction, and decouple the mitigation process into a preference optimization problem. Furthermore, we construct "CP-Bench", a novel benchmark to evaluate LVLMs' ability to correctly handle CPQs and produce factual responses. Applied to the LLaVA series, Antidote can simultaneously enhance performance on CP-Bench by over 50%, POPE by 1.8-3.3%, and CHAIR & SHR by 30-50%, all without relying on external supervision from stronger LVLMs or human feedback and introducing noticeable catastrophic forgetting issues.
HarmonySeg: Tubular Structure Segmentation with Deep-Shallow Feature Fusion and Growth-Suppression Balanced Loss
Apr 10, 2025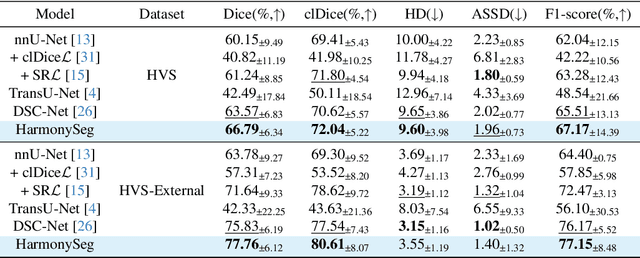
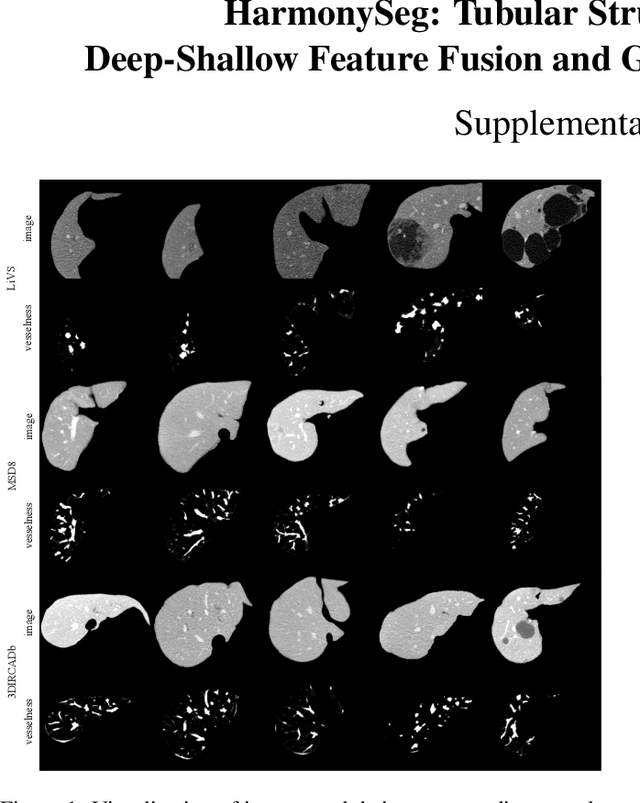
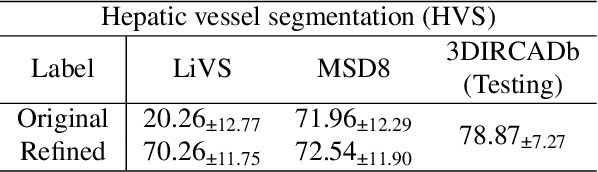
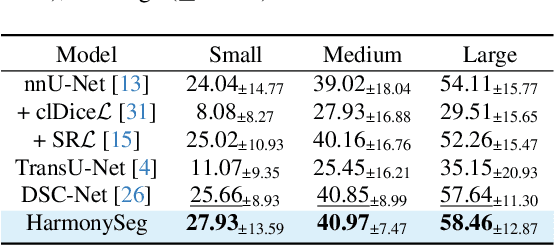
Abstract:Accurate segmentation of tubular structures in medical images, such as vessels and airway trees, is crucial for computer-aided diagnosis, radiotherapy, and surgical planning. However, significant challenges exist in algorithm design when faced with diverse sizes, complex topologies, and (often) incomplete data annotation of these structures. We address these difficulties by proposing a new tubular structure segmentation framework named HarmonySeg. First, we design a deep-to-shallow decoder network featuring flexible convolution blocks with varying receptive fields, which enables the model to effectively adapt to tubular structures of different scales. Second, to highlight potential anatomical regions and improve the recall of small tubular structures, we incorporate vesselness maps as auxiliary information. These maps are aligned with image features through a shallow-and-deep fusion module, which simultaneously eliminates unreasonable candidates to maintain high precision. Finally, we introduce a topology-preserving loss function that leverages contextual and shape priors to balance the growth and suppression of tubular structures, which also allows the model to handle low-quality and incomplete annotations. Extensive quantitative experiments are conducted on four public datasets. The results show that our model can accurately segment 2D and 3D tubular structures and outperform existing state-of-the-art methods. External validation on a private dataset also demonstrates good generalizability.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge