Yun Bian
MMSF: Multitask and Multimodal Supervised Framework for WSI Classification and Survival Analysis
Jan 28, 2026Abstract:Multimodal evidence is critical in computational pathology: gigapixel whole slide images capture tumor morphology, while patient-level clinical descriptors preserve complementary context for prognosis. Integrating such heterogeneous signals remains challenging because feature spaces exhibit distinct statistics and scales. We introduce MMSF, a multitask and multimodal supervised framework built on a linear-complexity MIL backbone that explicitly decomposes and fuses cross-modal information. MMSF comprises a graph feature extraction module embedding tissue topology at the patch level, a clinical data embedding module standardizing patient attributes, a feature fusion module aligning modality-shared and modality-specific representations, and a Mamba-based MIL encoder with multitask prediction heads. Experiments on CAMELYON16 and TCGA-NSCLC demonstrate 2.1--6.6\% accuracy and 2.2--6.9\% AUC improvements over competitive baselines, while evaluations on five TCGA survival cohorts yield 7.1--9.8\% C-index improvements compared with unimodal methods and 5.6--7.1\% over multimodal alternatives.
Focus on What Matters: Fisher-Guided Adaptive Multimodal Fusion for Vulnerability Detection
Jan 05, 2026Abstract:Software vulnerability detection is a critical task for securing software systems and can be formulated as a binary classification problem: given a code snippet, determine whether it contains a vulnerability. Existing multimodal approaches typically fuse Natural Code Sequence (NCS) representations from pretrained language models with Code Property Graph (CPG) representations from graph neural networks, often under the implicit assumption that adding a modality necessarily yields extra information. In practice, sequence and graph representations can be redundant, and fluctuations in the quality of the graph modality can dilute the discriminative signal of the dominant modality. To address this, we propose TaCCS-DFA, a framework that introduces Fisher information as a geometric measure of how sensitive feature directions are to the classification decision, enabling task-oriented complementary fusion. TaCCS-DFA online estimates a low-rank principal Fisher subspace and restricts cross-modal attention to task-sensitive directions, thereby retrieving structural features from CPG that complement the sequence modality; meanwhile, an adaptive gating mechanism dynamically adjusts the contribution of the graph modality for each sample to suppress noise propagation. Our analysis shows that, under an isotropic perturbation assumption, the proposed mechanism admits a tighter risk bound than conventional full-spectrum attention. Experiments on BigVul, Devign, and ReVeal show that TaCCS-DFA achieves strong performance across multiple backbones. With CodeT5 as the backbone, TaCCS-DFA reaches an F1 score of 87.80\% on the highly imbalanced BigVul dataset, improving over a strong baseline Vul-LMGNNs by 6.3 percentage points while maintaining low calibration error and computational overhead.
MUSE: Multi-Scale Dense Self-Distillation for Nucleus Detection and Classification
Nov 07, 2025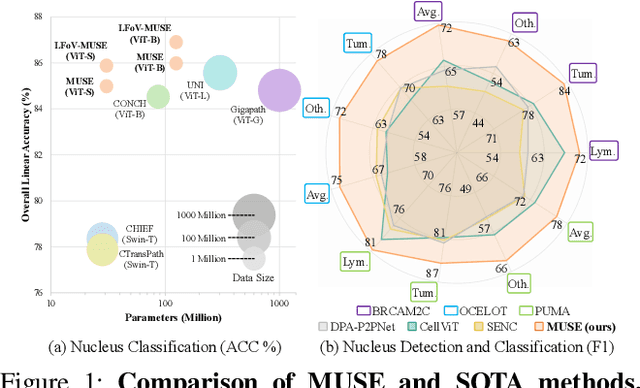
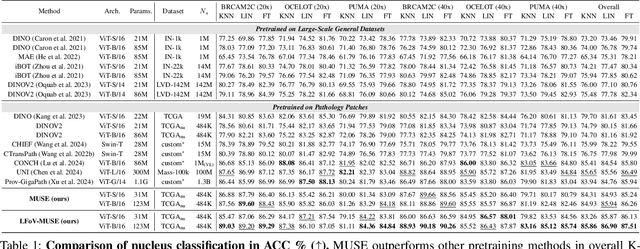
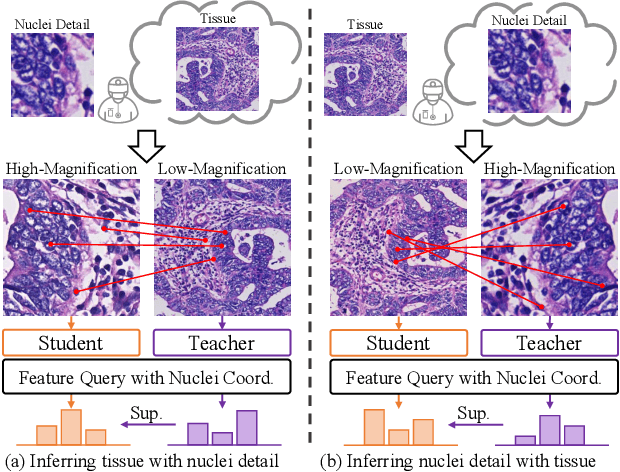
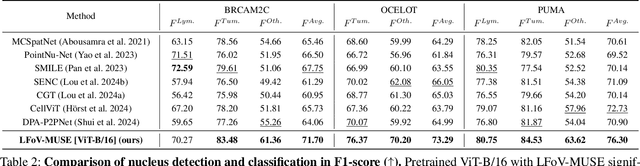
Abstract:Nucleus detection and classification (NDC) in histopathology analysis is a fundamental task that underpins a wide range of high-level pathology applications. However, existing methods heavily rely on labor-intensive nucleus-level annotations and struggle to fully exploit large-scale unlabeled data for learning discriminative nucleus representations. In this work, we propose MUSE (MUlti-scale denSE self-distillation), a novel self-supervised learning method tailored for NDC. At its core is NuLo (Nucleus-based Local self-distillation), a coordinate-guided mechanism that enables flexible local self-distillation based on predicted nucleus positions. By removing the need for strict spatial alignment between augmented views, NuLo allows critical cross-scale alignment, thus unlocking the capacity of models for fine-grained nucleus-level representation. To support MUSE, we design a simple yet effective encoder-decoder architecture and a large field-of-view semi-supervised fine-tuning strategy that together maximize the value of unlabeled pathology images. Extensive experiments on three widely used benchmarks demonstrate that MUSE effectively addresses the core challenges of histopathological NDC. The resulting models not only surpass state-of-the-art supervised baselines but also outperform generic pathology foundation models.
A Continual Learning-driven Model for Accurate and Generalizable Segmentation of Clinically Comprehensive and Fine-grained Whole-body Anatomies in CT
Mar 16, 2025Abstract:Precision medicine in the quantitative management of chronic diseases and oncology would be greatly improved if the Computed Tomography (CT) scan of any patient could be segmented, parsed and analyzed in a precise and detailed way. However, there is no such fully annotated CT dataset with all anatomies delineated for training because of the exceptionally high manual cost, the need for specialized clinical expertise, and the time required to finish the task. To this end, we proposed a novel continual learning-driven CT model that can segment complete anatomies presented using dozens of previously partially labeled datasets, dynamically expanding its capacity to segment new ones without compromising previously learned organ knowledge. Existing multi-dataset approaches are not able to dynamically segment new anatomies without catastrophic forgetting and would encounter optimization difficulty or infeasibility when segmenting hundreds of anatomies across the whole range of body regions. Our single unified CT segmentation model, CL-Net, can highly accurately segment a clinically comprehensive set of 235 fine-grained whole-body anatomies. Composed of a universal encoder, multiple optimized and pruned decoders, CL-Net is developed using 13,952 CT scans from 20 public and 16 private high-quality partially labeled CT datasets of various vendors, different contrast phases, and pathologies. Extensive evaluation demonstrates that CL-Net consistently outperforms the upper limit of an ensemble of 36 specialist nnUNets trained per dataset with the complexity of 5% model size and significantly surpasses the segmentation accuracy of recent leading Segment Anything-style medical image foundation models by large margins. Our continual learning-driven CL-Net model would lay a solid foundation to facilitate many downstream tasks of oncology and chronic diseases using the most widely adopted CT imaging.
From Slices to Sequences: Autoregressive Tracking Transformer for Cohesive and Consistent 3D Lymph Node Detection in CT Scans
Mar 11, 2025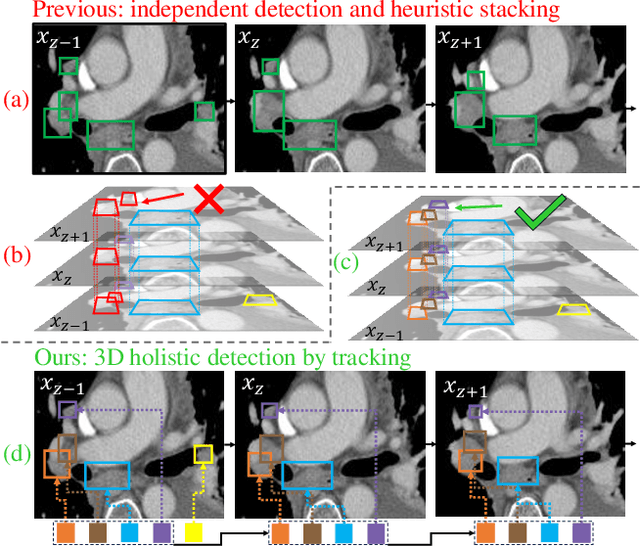

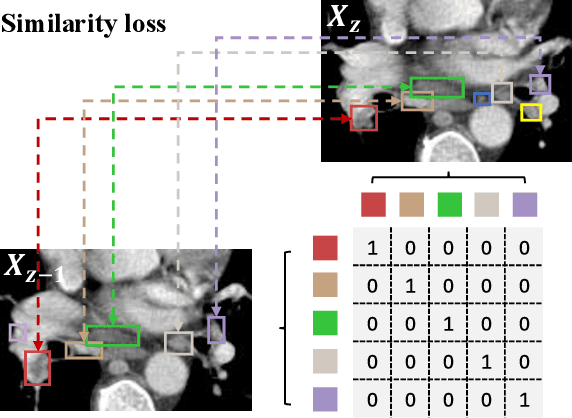
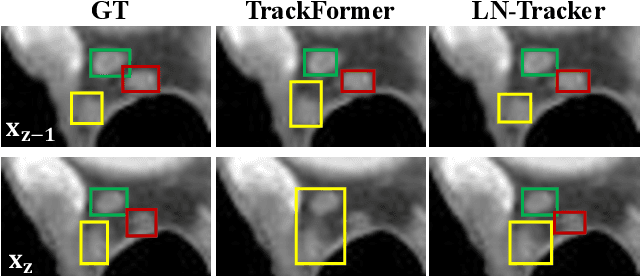
Abstract:Lymph node (LN) assessment is an essential task in the routine radiology workflow, providing valuable insights for cancer staging, treatment planning and beyond. Identifying scatteredly-distributed and low-contrast LNs in 3D CT scans is highly challenging, even for experienced clinicians. Previous lesion and LN detection methods demonstrate effectiveness of 2.5D approaches (i.e, using 2D network with multi-slice inputs), leveraging pretrained 2D model weights and showing improved accuracy as compared to separate 2D or 3D detectors. However, slice-based 2.5D detectors do not explicitly model inter-slice consistency for LN as a 3D object, requiring heuristic post-merging steps to generate final 3D LN instances, which can involve tuning a set of parameters for each dataset. In this work, we formulate 3D LN detection as a tracking task and propose LN-Tracker, a novel LN tracking transformer, for joint end-to-end detection and 3D instance association. Built upon DETR-based detector, LN-Tracker decouples transformer decoder's query into the track and detection groups, where the track query autoregressively follows previously tracked LN instances along the z-axis of a CT scan. We design a new transformer decoder with masked attention module to align track query's content to the context of current slice, meanwhile preserving detection query's high accuracy in current slice. An inter-slice similarity loss is introduced to encourage cohesive LN association between slices. Extensive evaluation on four lymph node datasets shows LN-Tracker's superior performance, with at least 2.7% gain in average sensitivity when compared to other top 3D/2.5D detectors. Further validation on public lung nodule and prostate tumor detection tasks confirms the generalizability of LN-Tracker as it achieves top performance on both tasks. Datasets will be released upon acceptance.
From Histopathology Images to Cell Clouds: Learning Slide Representations with Hierarchical Cell Transformer
Dec 21, 2024Abstract:It is clinically crucial and potentially very beneficial to be able to analyze and model directly the spatial distributions of cells in histopathology whole slide images (WSI). However, most existing WSI datasets lack cell-level annotations, owing to the extremely high cost over giga-pixel images. Thus, it remains an open question whether deep learning models can directly and effectively analyze WSIs from the semantic aspect of cell distributions. In this work, we construct a large-scale WSI dataset with more than 5 billion cell-level annotations, termed WSI-Cell5B, and a novel hierarchical Cell Cloud Transformer (CCFormer) to tackle these challenges. WSI-Cell5B is based on 6,998 WSIs of 11 cancers from The Cancer Genome Atlas Program, and all WSIs are annotated per cell by coordinates and types. To the best of our knowledge, WSI-Cell5B is the first WSI-level large-scale dataset integrating cell-level annotations. On the other hand, CCFormer formulates the collection of cells in each WSI as a cell cloud and models cell spatial distribution. Specifically, Neighboring Information Embedding (NIE) is proposed to characterize the distribution of cells within the neighborhood of each cell, and a novel Hierarchical Spatial Perception (HSP) module is proposed to learn the spatial relationship among cells in a bottom-up manner. The clinical analysis indicates that WSI-Cell5B can be used to design clinical evaluation metrics based on counting cells that effectively assess the survival risk of patients. Extensive experiments on survival prediction and cancer staging show that learning from cell spatial distribution alone can already achieve state-of-the-art (SOTA) performance, i.e., CCFormer strongly outperforms other competing methods.
From Pixels to Gigapixels: Bridging Local Inductive Bias and Long-Range Dependencies with Pixel-Mamba
Dec 21, 2024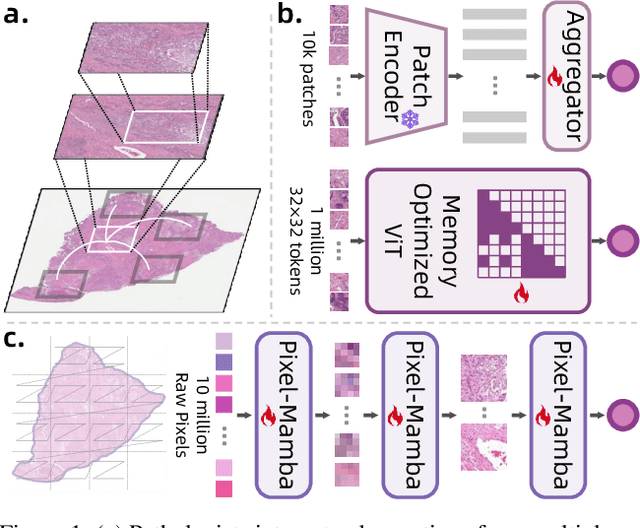
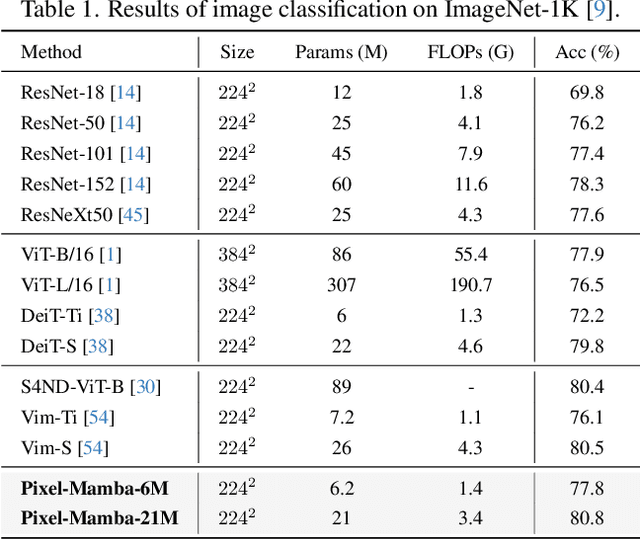
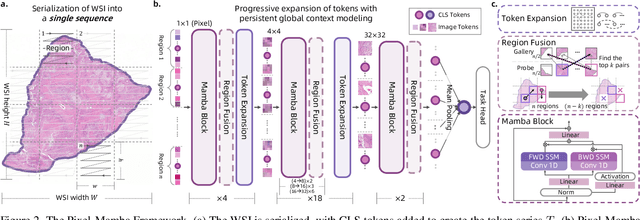
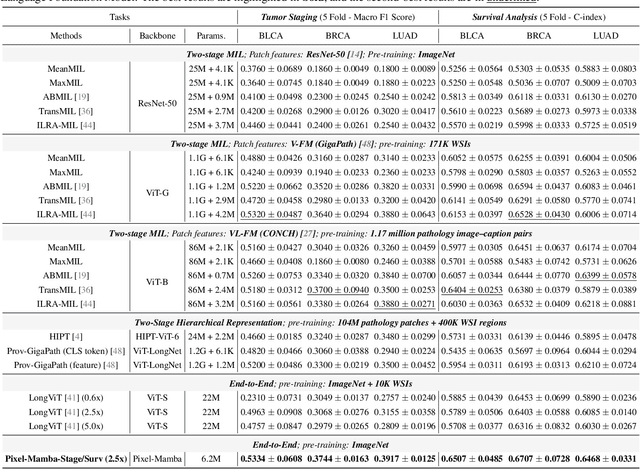
Abstract:Histopathology plays a critical role in medical diagnostics, with whole slide images (WSIs) offering valuable insights that directly influence clinical decision-making. However, the large size and complexity of WSIs may pose significant challenges for deep learning models, in both computational efficiency and effective representation learning. In this work, we introduce Pixel-Mamba, a novel deep learning architecture designed to efficiently handle gigapixel WSIs. Pixel-Mamba leverages the Mamba module, a state-space model (SSM) with linear memory complexity, and incorporates local inductive biases through progressively expanding tokens, akin to convolutional neural networks. This enables Pixel-Mamba to hierarchically combine both local and global information while efficiently addressing computational challenges. Remarkably, Pixel-Mamba achieves or even surpasses the quantitative performance of state-of-the-art (SOTA) foundation models that were pretrained on millions of WSIs or WSI-text pairs, in a range of tumor staging and survival analysis tasks, {\bf even without requiring any pathology-specific pretraining}. Extensive experiments demonstrate the efficacy of Pixel-Mamba as a powerful and efficient framework for end-to-end WSI analysis.
End-to-end Multi-source Visual Prompt Tuning for Survival Analysis in Whole Slide Images
Sep 05, 2024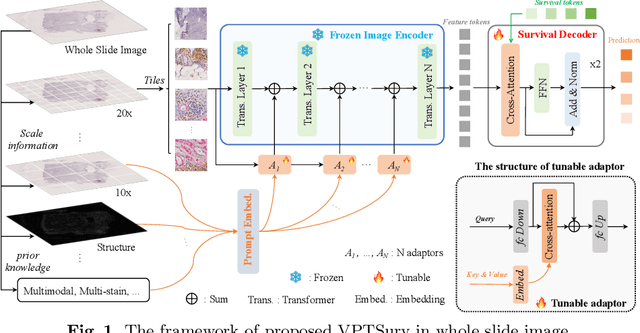
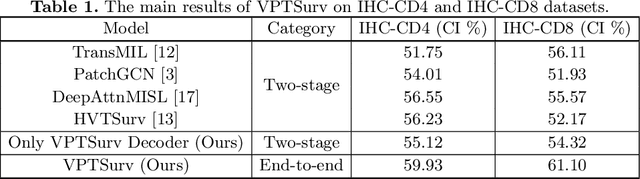
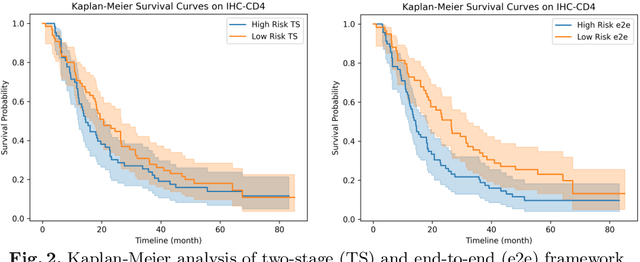
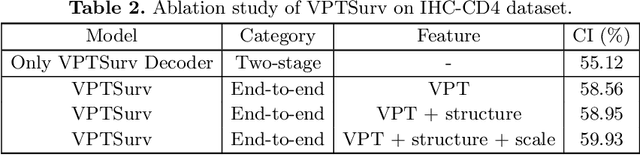
Abstract:Survival analysis using pathology images poses a considerable challenge, as it requires the localization of relevant information from the multitude of tiles within whole slide images (WSIs). Current methods typically resort to a two-stage approach, where a pre-trained network extracts features from tiles, which are then used by survival models. This process, however, does not optimize the survival models in an end-to-end manner, and the pre-extracted features may not be ideally suited for survival prediction. To address this limitation, we present a novel end-to-end Visual Prompt Tuning framework for survival analysis, named VPTSurv. VPTSurv refines feature embeddings through an efficient encoder-decoder framework. The encoder remains fixed while the framework introduces tunable visual prompts and adaptors, thus permitting end-to-end training specifically for survival prediction by optimizing only the lightweight adaptors and the decoder. Moreover, the versatile VPTSurv framework accommodates multi-source information as prompts, thereby enriching the survival model. VPTSurv achieves substantial increases of 8.7% and 12.5% in the C-index on two immunohistochemical pathology image datasets. These significant improvements highlight the transformative potential of the end-to-end VPT framework over traditional two-stage methods.
A deep local attention network for pre-operative lymph node metastasis prediction in pancreatic cancer via multiphase CT imaging
Jan 04, 2023



Abstract:Lymph node (LN) metastasis status is one of the most critical prognostic and cancer staging factors for patients with resectable pancreatic ductal adenocarcinoma (PDAC), or in general, for any types of solid malignant tumors. Preoperative prediction of LN metastasis from non-invasive CT imaging is highly desired, as it might be straightforwardly used to guide the following neoadjuvant treatment decision and surgical planning. Most studies only capture the tumor characteristics in CT imaging to implicitly infer LN metastasis and very few work exploit direct LN's CT imaging information. To the best of our knowledge, this is the first work to propose a fully-automated LN segmentation and identification network to directly facilitate the LN metastasis status prediction task. Nevertheless LN segmentation/detection is very challenging since LN can be easily confused with other hard negative anatomic structures (e.g., vessels) from radiological images. We explore the anatomical spatial context priors of pancreatic LN locations by generating a guiding attention map from related organs and vessels to assist segmentation and infer LN status. As such, LN segmentation is impelled to focus on regions that are anatomically adjacent or plausible with respect to the specific organs and vessels. The metastasized LN identification network is trained to classify the segmented LN instances into positives or negatives by reusing the segmentation network as a pre-trained backbone and padding a new classification head. More importantly, we develop a LN metastasis status prediction network that combines the patient-wise aggregation results of LN segmentation/identification and deep imaging features extracted from the tumor region. Extensive quantitative nested five-fold cross-validation is conducted on a discovery dataset of 749 patients with PDAC.
Robust Pancreatic Ductal Adenocarcinoma Segmentation with Multi-Institutional Multi-Phase Partially-Annotated CT Scans
Aug 24, 2020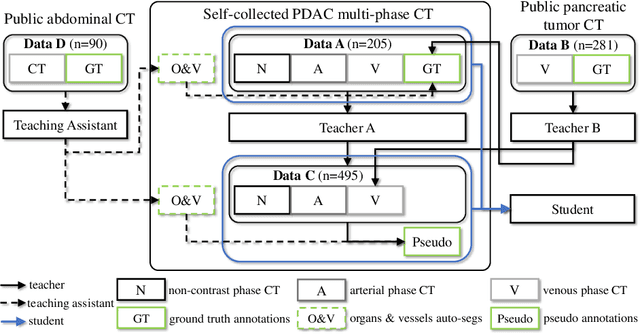
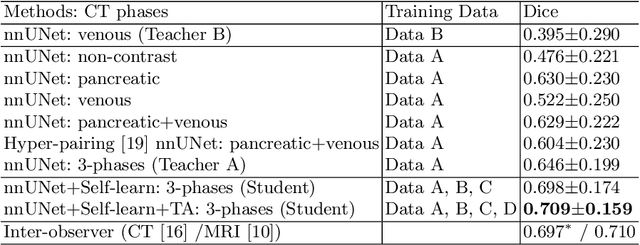
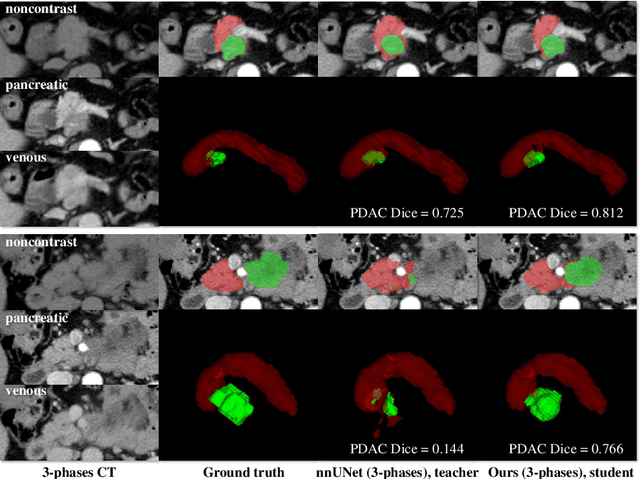
Abstract:Accurate and automated tumor segmentation is highly desired since it has the great potential to increase the efficiency and reproducibility of computing more complete tumor measurements and imaging biomarkers, comparing to (often partial) human measurements. This is probably the only viable means to enable the large-scale clinical oncology patient studies that utilize medical imaging. Deep learning approaches have shown robust segmentation performances for certain types of tumors, e.g., brain tumors in MRI imaging, when a training dataset with plenty of pixel-level fully-annotated tumor images is available. However, more than often, we are facing the challenge that only (very) limited annotations are feasible to acquire, especially for hard tumors. Pancreatic ductal adenocarcinoma (PDAC) segmentation is one of the most challenging tumor segmentation tasks, yet critically important for clinical needs. Previous work on PDAC segmentation is limited to the moderate amounts of annotated patient images (n<300) from venous or venous+arterial phase CT scans. Based on a new self-learning framework, we propose to train the PDAC segmentation model using a much larger quantity of patients (n~=1,000), with a mix of annotated and un-annotated venous or multi-phase CT images. Pseudo annotations are generated by combining two teacher models with different PDAC segmentation specialties on unannotated images, and can be further refined by a teaching assistant model that identifies associated vessels around the pancreas. A student model is trained on both manual and pseudo annotated multi-phase images. Experiment results show that our proposed method provides an absolute improvement of 6.3% Dice score over the strong baseline of nnUNet trained on annotated images, achieving the performance (Dice = 0.71) similar to the inter-observer variability between radiologists.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge