Le Lu
Non-Contrast CT Esophageal Varices Grading through Clinical Prior-Enhanced Multi-Organ Analysis
Dec 22, 2025Abstract:Esophageal varices (EV) represent a critical complication of portal hypertension, affecting approximately 60% of cirrhosis patients with a significant bleeding risk of ~30%. While traditionally diagnosed through invasive endoscopy, non-contrast computed tomography (NCCT) presents a potential non-invasive alternative that has yet to be fully utilized in clinical practice. We present Multi-Organ-COhesion Network++ (MOON++), a novel multimodal framework that enhances EV assessment through comprehensive analysis of NCCT scans. Inspired by clinical evidence correlating organ volumetric relationships with liver disease severity, MOON++ synthesizes imaging characteristics of the esophagus, liver, and spleen through multimodal learning. We evaluated our approach using 1,631 patients, those with endoscopically confirmed EV were classified into four severity grades. Validation in 239 patient cases and independent testing in 289 cases demonstrate superior performance compared to conventional single organ methods, achieving an AUC of 0.894 versus 0.803 for the severe grade EV classification (G3 versus <G3) and 0.921 versus 0.793 for the differentiation of moderate to severe grades (>=G2 versus <G2). We conducted a reader study involving experienced radiologists to further validate the performance of MOON++. To our knowledge, MOON++ represents the first comprehensive multi-organ NCCT analysis framework incorporating clinical knowledge priors for EV assessment, potentially offering a promising non-invasive diagnostic alternative.
MUSE: Multi-Scale Dense Self-Distillation for Nucleus Detection and Classification
Nov 07, 2025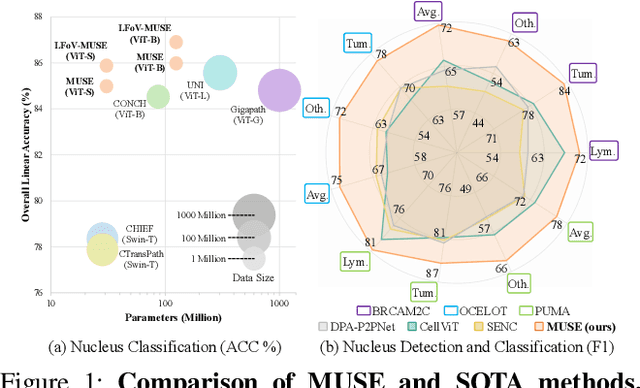
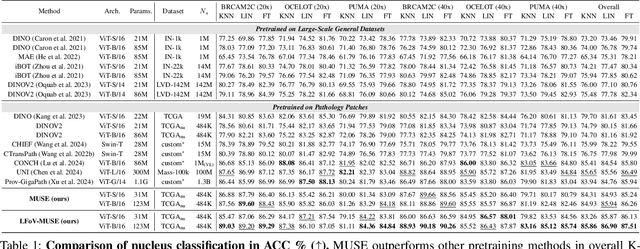
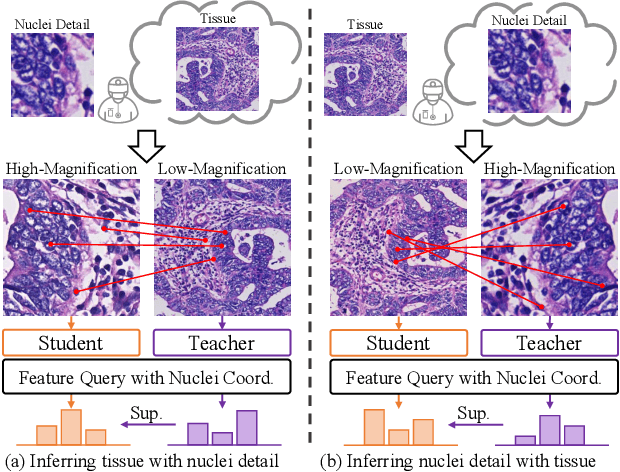
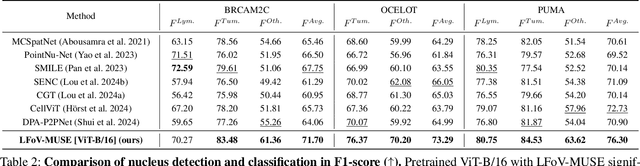
Abstract:Nucleus detection and classification (NDC) in histopathology analysis is a fundamental task that underpins a wide range of high-level pathology applications. However, existing methods heavily rely on labor-intensive nucleus-level annotations and struggle to fully exploit large-scale unlabeled data for learning discriminative nucleus representations. In this work, we propose MUSE (MUlti-scale denSE self-distillation), a novel self-supervised learning method tailored for NDC. At its core is NuLo (Nucleus-based Local self-distillation), a coordinate-guided mechanism that enables flexible local self-distillation based on predicted nucleus positions. By removing the need for strict spatial alignment between augmented views, NuLo allows critical cross-scale alignment, thus unlocking the capacity of models for fine-grained nucleus-level representation. To support MUSE, we design a simple yet effective encoder-decoder architecture and a large field-of-view semi-supervised fine-tuning strategy that together maximize the value of unlabeled pathology images. Extensive experiments on three widely used benchmarks demonstrate that MUSE effectively addresses the core challenges of histopathological NDC. The resulting models not only surpass state-of-the-art supervised baselines but also outperform generic pathology foundation models.
Anatomy-Aware Low-Dose CT Denoising via Pretrained Vision Models and Semantic-Guided Contrastive Learning
Aug 11, 2025Abstract:To reduce radiation exposure and improve the diagnostic efficacy of low-dose computed tomography (LDCT), numerous deep learning-based denoising methods have been developed to mitigate noise and artifacts. However, most of these approaches ignore the anatomical semantics of human tissues, which may potentially result in suboptimal denoising outcomes. To address this problem, we propose ALDEN, an anatomy-aware LDCT denoising method that integrates semantic features of pretrained vision models (PVMs) with adversarial and contrastive learning. Specifically, we introduce an anatomy-aware discriminator that dynamically fuses hierarchical semantic features from reference normal-dose CT (NDCT) via cross-attention mechanisms, enabling tissue-specific realism evaluation in the discriminator. In addition, we propose a semantic-guided contrastive learning module that enforces anatomical consistency by contrasting PVM-derived features from LDCT, denoised CT and NDCT, preserving tissue-specific patterns through positive pairs and suppressing artifacts via dual negative pairs. Extensive experiments conducted on two LDCT denoising datasets reveal that ALDEN achieves the state-of-the-art performance, offering superior anatomy preservation and substantially reducing over-smoothing issue of previous work. Further validation on a downstream multi-organ segmentation task (encompassing 117 anatomical structures) affirms the model's ability to maintain anatomical awareness.
HarmonySeg: Tubular Structure Segmentation with Deep-Shallow Feature Fusion and Growth-Suppression Balanced Loss
Apr 10, 2025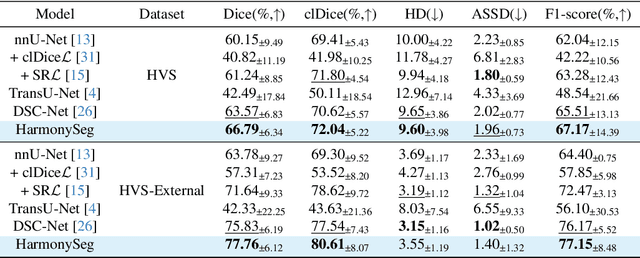
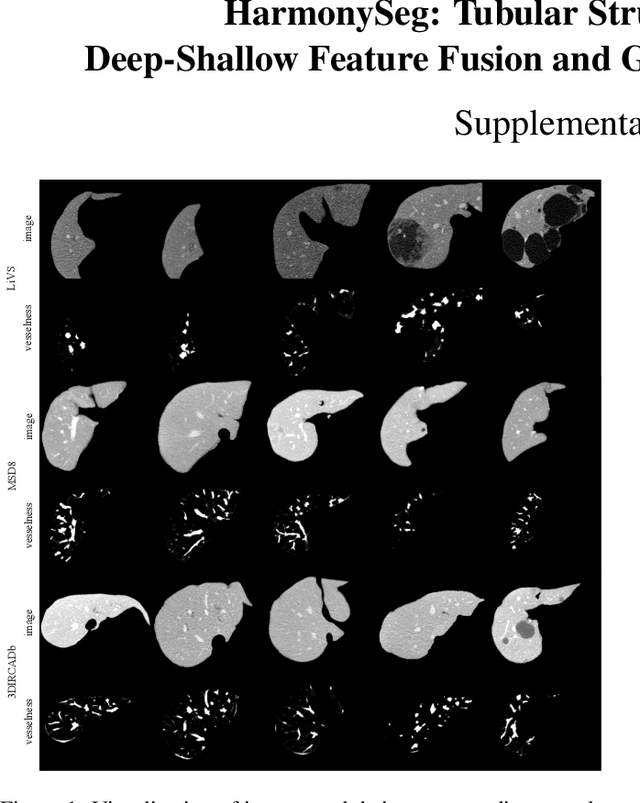
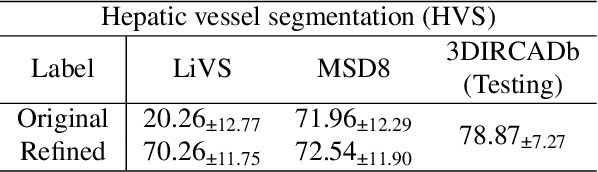
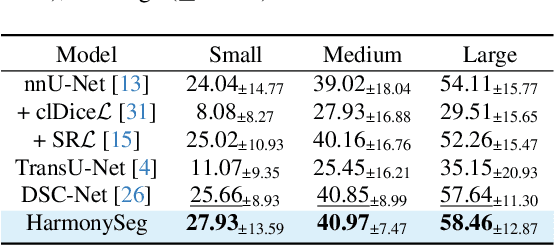
Abstract:Accurate segmentation of tubular structures in medical images, such as vessels and airway trees, is crucial for computer-aided diagnosis, radiotherapy, and surgical planning. However, significant challenges exist in algorithm design when faced with diverse sizes, complex topologies, and (often) incomplete data annotation of these structures. We address these difficulties by proposing a new tubular structure segmentation framework named HarmonySeg. First, we design a deep-to-shallow decoder network featuring flexible convolution blocks with varying receptive fields, which enables the model to effectively adapt to tubular structures of different scales. Second, to highlight potential anatomical regions and improve the recall of small tubular structures, we incorporate vesselness maps as auxiliary information. These maps are aligned with image features through a shallow-and-deep fusion module, which simultaneously eliminates unreasonable candidates to maintain high precision. Finally, we introduce a topology-preserving loss function that leverages contextual and shape priors to balance the growth and suppression of tubular structures, which also allows the model to handle low-quality and incomplete annotations. Extensive quantitative experiments are conducted on four public datasets. The results show that our model can accurately segment 2D and 3D tubular structures and outperform existing state-of-the-art methods. External validation on a private dataset also demonstrates good generalizability.
Mamba-3D as Masked Autoencoders for Accurate and Data-Efficient Analysis of Medical Ultrasound Videos
Mar 26, 2025Abstract:Ultrasound videos are an important form of clinical imaging data, and deep learning-based automated analysis can improve diagnostic accuracy and clinical efficiency. However, the scarcity of labeled data and the inherent challenges of video analysis have impeded the advancement of related methods. In this work, we introduce E-ViM$^3$, a data-efficient Vision Mamba network that preserves the 3D structure of video data, enhancing long-range dependencies and inductive biases to better model space-time correlations. With our design of Enclosure Global Tokens (EGT), the model captures and aggregates global features more effectively than competing methods. To further improve data efficiency, we employ masked video modeling for self-supervised pre-training, with the proposed Spatial-Temporal Chained (STC) masking strategy designed to adapt to various video scenarios. Experiments demonstrate that E-ViM$^3$ performs as the state-of-the-art in two high-level semantic analysis tasks across four datasets of varying sizes: EchoNet-Dynamic, CAMUS, MICCAI-BUV, and WHBUS. Furthermore, our model achieves competitive performance with limited labels, highlighting its potential impact on real-world clinical applications.
UniReg: Foundation Model for Controllable Medical Image Registration
Mar 17, 2025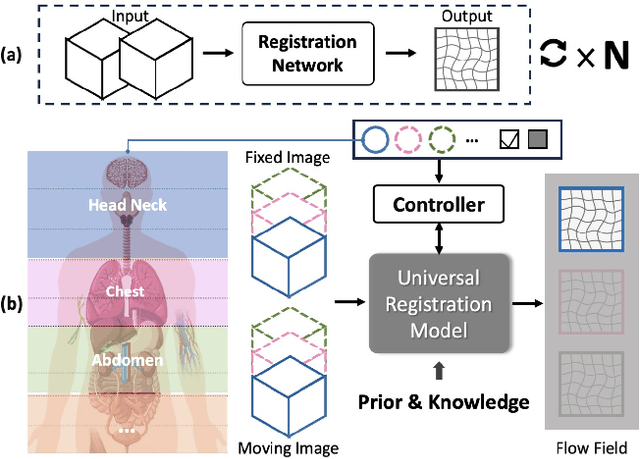
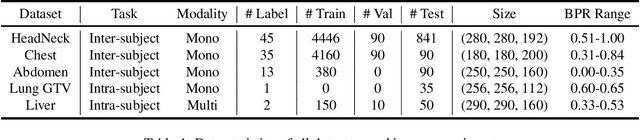
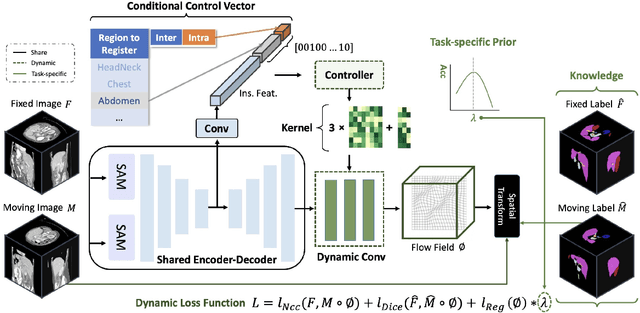
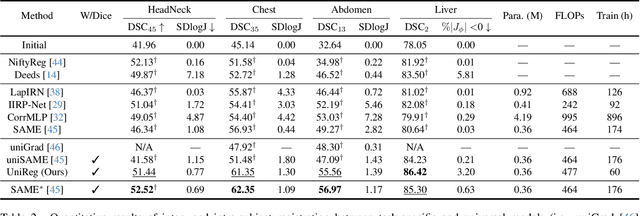
Abstract:Learning-based medical image registration has achieved performance parity with conventional methods while demonstrating a substantial advantage in computational efficiency. However, learning-based registration approaches lack generalizability across diverse clinical scenarios, requiring the laborious development of multiple isolated networks for specific registration tasks, e.g., inter-/intra-subject registration or organ-specific alignment. % To overcome this limitation, we propose \textbf{UniReg}, the first interactive foundation model for medical image registration, which combines the precision advantages of task-specific learning methods with the generalization of traditional optimization methods. Our key innovation is a unified framework for diverse registration scenarios, achieved through a conditional deformation field estimation within a unified registration model. This is realized through a dynamic learning paradigm that explicitly encodes: (1) anatomical structure priors, (2) registration type constraints (inter/intra-subject), and (3) instance-specific features, enabling the generation of scenario-optimal deformation fields. % Through comprehensive experiments encompassing $90$ anatomical structures at different body regions, our UniReg model demonstrates comparable performance with contemporary state-of-the-art methodologies while achieving ~50\% reduction in required training iterations relative to the conventional learning-based paradigm. This optimization contributes to a significant reduction in computational resources, such as training time. Code and model will be available.
A Continual Learning-driven Model for Accurate and Generalizable Segmentation of Clinically Comprehensive and Fine-grained Whole-body Anatomies in CT
Mar 16, 2025Abstract:Precision medicine in the quantitative management of chronic diseases and oncology would be greatly improved if the Computed Tomography (CT) scan of any patient could be segmented, parsed and analyzed in a precise and detailed way. However, there is no such fully annotated CT dataset with all anatomies delineated for training because of the exceptionally high manual cost, the need for specialized clinical expertise, and the time required to finish the task. To this end, we proposed a novel continual learning-driven CT model that can segment complete anatomies presented using dozens of previously partially labeled datasets, dynamically expanding its capacity to segment new ones without compromising previously learned organ knowledge. Existing multi-dataset approaches are not able to dynamically segment new anatomies without catastrophic forgetting and would encounter optimization difficulty or infeasibility when segmenting hundreds of anatomies across the whole range of body regions. Our single unified CT segmentation model, CL-Net, can highly accurately segment a clinically comprehensive set of 235 fine-grained whole-body anatomies. Composed of a universal encoder, multiple optimized and pruned decoders, CL-Net is developed using 13,952 CT scans from 20 public and 16 private high-quality partially labeled CT datasets of various vendors, different contrast phases, and pathologies. Extensive evaluation demonstrates that CL-Net consistently outperforms the upper limit of an ensemble of 36 specialist nnUNets trained per dataset with the complexity of 5% model size and significantly surpasses the segmentation accuracy of recent leading Segment Anything-style medical image foundation models by large margins. Our continual learning-driven CL-Net model would lay a solid foundation to facilitate many downstream tasks of oncology and chronic diseases using the most widely adopted CT imaging.
From Slices to Sequences: Autoregressive Tracking Transformer for Cohesive and Consistent 3D Lymph Node Detection in CT Scans
Mar 11, 2025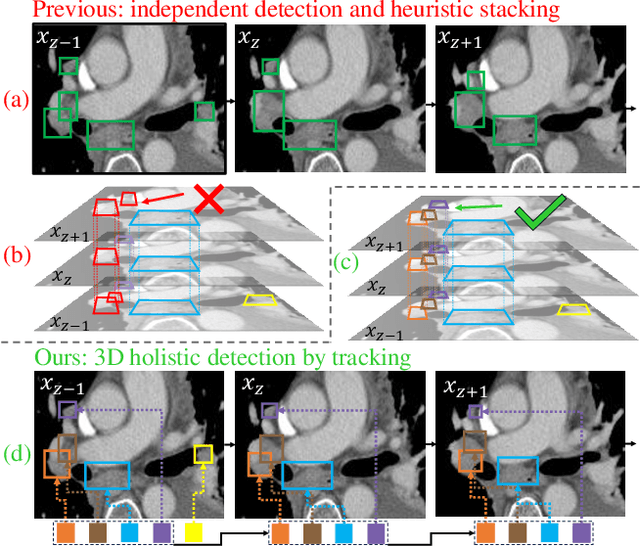

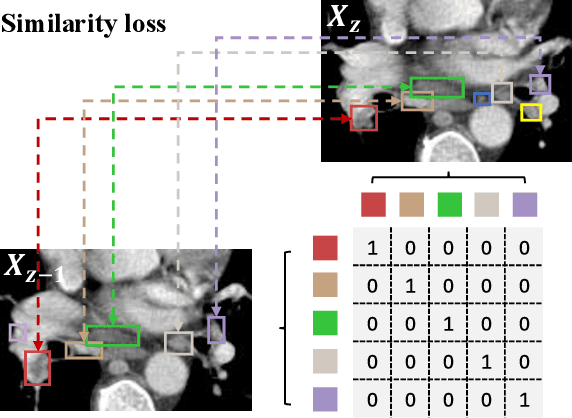
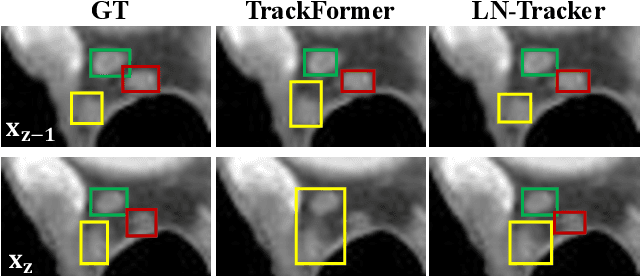
Abstract:Lymph node (LN) assessment is an essential task in the routine radiology workflow, providing valuable insights for cancer staging, treatment planning and beyond. Identifying scatteredly-distributed and low-contrast LNs in 3D CT scans is highly challenging, even for experienced clinicians. Previous lesion and LN detection methods demonstrate effectiveness of 2.5D approaches (i.e, using 2D network with multi-slice inputs), leveraging pretrained 2D model weights and showing improved accuracy as compared to separate 2D or 3D detectors. However, slice-based 2.5D detectors do not explicitly model inter-slice consistency for LN as a 3D object, requiring heuristic post-merging steps to generate final 3D LN instances, which can involve tuning a set of parameters for each dataset. In this work, we formulate 3D LN detection as a tracking task and propose LN-Tracker, a novel LN tracking transformer, for joint end-to-end detection and 3D instance association. Built upon DETR-based detector, LN-Tracker decouples transformer decoder's query into the track and detection groups, where the track query autoregressively follows previously tracked LN instances along the z-axis of a CT scan. We design a new transformer decoder with masked attention module to align track query's content to the context of current slice, meanwhile preserving detection query's high accuracy in current slice. An inter-slice similarity loss is introduced to encourage cohesive LN association between slices. Extensive evaluation on four lymph node datasets shows LN-Tracker's superior performance, with at least 2.7% gain in average sensitivity when compared to other top 3D/2.5D detectors. Further validation on public lung nodule and prostate tumor detection tasks confirms the generalizability of LN-Tracker as it achieves top performance on both tasks. Datasets will be released upon acceptance.
MRS: A Fast Sampler for Mean Reverting Diffusion based on ODE and SDE Solvers
Feb 11, 2025Abstract:In applications of diffusion models, controllable generation is of practical significance, but is also challenging. Current methods for controllable generation primarily focus on modifying the score function of diffusion models, while Mean Reverting (MR) Diffusion directly modifies the structure of the stochastic differential equation (SDE), making the incorporation of image conditions simpler and more natural. However, current training-free fast samplers are not directly applicable to MR Diffusion. And thus MR Diffusion requires hundreds of NFEs (number of function evaluations) to obtain high-quality samples. In this paper, we propose a new algorithm named MRS (MR Sampler) to reduce the sampling NFEs of MR Diffusion. We solve the reverse-time SDE and the probability flow ordinary differential equation (PF-ODE) associated with MR Diffusion, and derive semi-analytical solutions. The solutions consist of an analytical function and an integral parameterized by a neural network. Based on this solution, we can generate high-quality samples in fewer steps. Our approach does not require training and supports all mainstream parameterizations, including noise prediction, data prediction and velocity prediction. Extensive experiments demonstrate that MR Sampler maintains high sampling quality with a speedup of 10 to 20 times across ten different image restoration tasks. Our algorithm accelerates the sampling procedure of MR Diffusion, making it more practical in controllable generation.
Large-scale and Fine-grained Vision-language Pre-training for Enhanced CT Image Understanding
Jan 24, 2025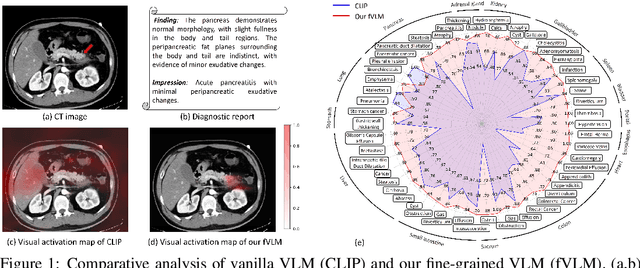
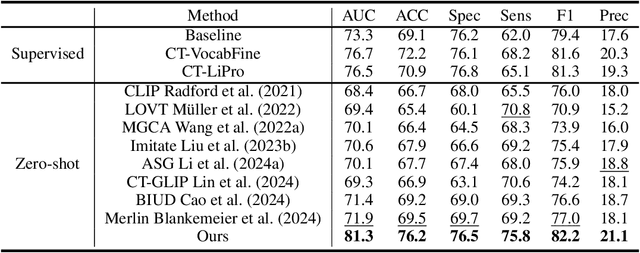
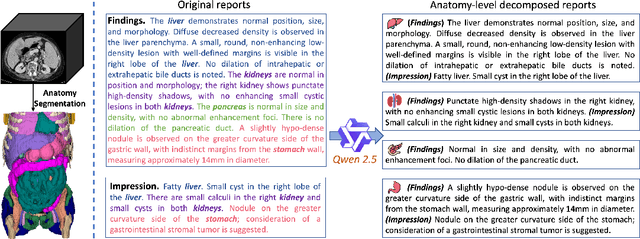
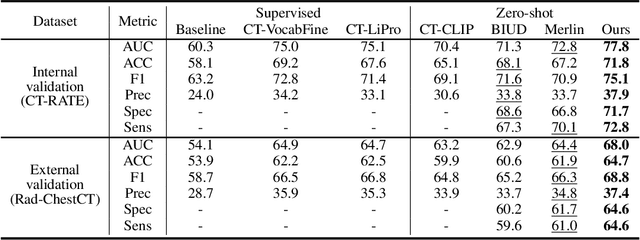
Abstract:Artificial intelligence (AI) shows great potential in assisting radiologists to improve the efficiency and accuracy of medical image interpretation and diagnosis. However, a versatile AI model requires large-scale data and comprehensive annotations, which are often impractical in medical settings. Recent studies leverage radiology reports as a naturally high-quality supervision for medical images, using contrastive language-image pre-training (CLIP) to develop language-informed models for radiological image interpretation. Nonetheless, these approaches typically contrast entire images with reports, neglecting the local associations between imaging regions and report sentences, which may undermine model performance and interoperability. In this paper, we propose a fine-grained vision-language model (fVLM) for anatomy-level CT image interpretation. Specifically, we explicitly match anatomical regions of CT images with corresponding descriptions in radiology reports and perform contrastive pre-training for each anatomy individually. Fine-grained alignment, however, faces considerable false-negative challenges, mainly from the abundance of anatomy-level healthy samples and similarly diseased abnormalities. To tackle this issue, we propose identifying false negatives of both normal and abnormal samples and calibrating contrastive learning from patient-level to disease-aware pairing. We curated the largest CT dataset to date, comprising imaging and report data from 69,086 patients, and conducted a comprehensive evaluation of 54 major and important disease diagnosis tasks across 15 main anatomies. Experimental results demonstrate the substantial potential of fVLM in versatile medical image interpretation. In the zero-shot classification task, we achieved an average AUC of 81.3% on 54 diagnosis tasks, surpassing CLIP and supervised methods by 12.9% and 8.0%, respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge