Qinji Yu
A Continual Learning-driven Model for Accurate and Generalizable Segmentation of Clinically Comprehensive and Fine-grained Whole-body Anatomies in CT
Mar 16, 2025Abstract:Precision medicine in the quantitative management of chronic diseases and oncology would be greatly improved if the Computed Tomography (CT) scan of any patient could be segmented, parsed and analyzed in a precise and detailed way. However, there is no such fully annotated CT dataset with all anatomies delineated for training because of the exceptionally high manual cost, the need for specialized clinical expertise, and the time required to finish the task. To this end, we proposed a novel continual learning-driven CT model that can segment complete anatomies presented using dozens of previously partially labeled datasets, dynamically expanding its capacity to segment new ones without compromising previously learned organ knowledge. Existing multi-dataset approaches are not able to dynamically segment new anatomies without catastrophic forgetting and would encounter optimization difficulty or infeasibility when segmenting hundreds of anatomies across the whole range of body regions. Our single unified CT segmentation model, CL-Net, can highly accurately segment a clinically comprehensive set of 235 fine-grained whole-body anatomies. Composed of a universal encoder, multiple optimized and pruned decoders, CL-Net is developed using 13,952 CT scans from 20 public and 16 private high-quality partially labeled CT datasets of various vendors, different contrast phases, and pathologies. Extensive evaluation demonstrates that CL-Net consistently outperforms the upper limit of an ensemble of 36 specialist nnUNets trained per dataset with the complexity of 5% model size and significantly surpasses the segmentation accuracy of recent leading Segment Anything-style medical image foundation models by large margins. Our continual learning-driven CL-Net model would lay a solid foundation to facilitate many downstream tasks of oncology and chronic diseases using the most widely adopted CT imaging.
From Slices to Sequences: Autoregressive Tracking Transformer for Cohesive and Consistent 3D Lymph Node Detection in CT Scans
Mar 11, 2025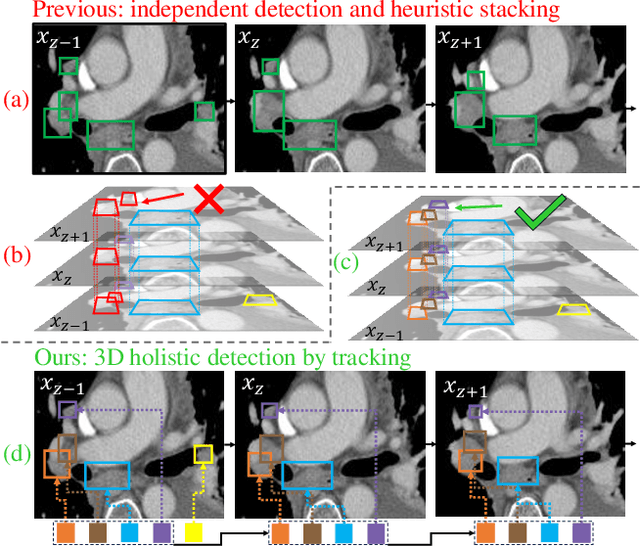

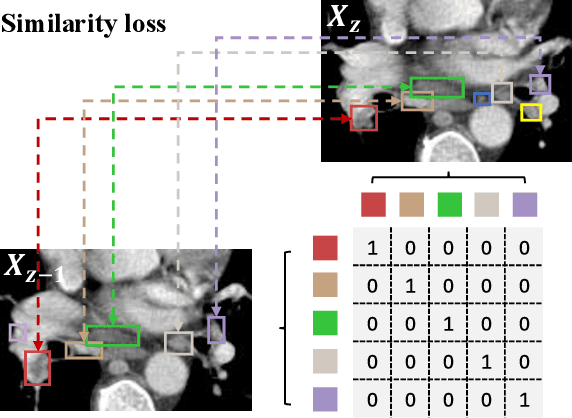
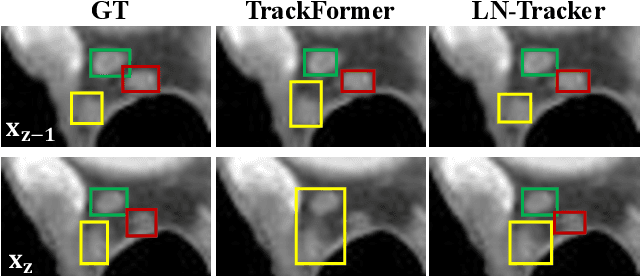
Abstract:Lymph node (LN) assessment is an essential task in the routine radiology workflow, providing valuable insights for cancer staging, treatment planning and beyond. Identifying scatteredly-distributed and low-contrast LNs in 3D CT scans is highly challenging, even for experienced clinicians. Previous lesion and LN detection methods demonstrate effectiveness of 2.5D approaches (i.e, using 2D network with multi-slice inputs), leveraging pretrained 2D model weights and showing improved accuracy as compared to separate 2D or 3D detectors. However, slice-based 2.5D detectors do not explicitly model inter-slice consistency for LN as a 3D object, requiring heuristic post-merging steps to generate final 3D LN instances, which can involve tuning a set of parameters for each dataset. In this work, we formulate 3D LN detection as a tracking task and propose LN-Tracker, a novel LN tracking transformer, for joint end-to-end detection and 3D instance association. Built upon DETR-based detector, LN-Tracker decouples transformer decoder's query into the track and detection groups, where the track query autoregressively follows previously tracked LN instances along the z-axis of a CT scan. We design a new transformer decoder with masked attention module to align track query's content to the context of current slice, meanwhile preserving detection query's high accuracy in current slice. An inter-slice similarity loss is introduced to encourage cohesive LN association between slices. Extensive evaluation on four lymph node datasets shows LN-Tracker's superior performance, with at least 2.7% gain in average sensitivity when compared to other top 3D/2.5D detectors. Further validation on public lung nodule and prostate tumor detection tasks confirms the generalizability of LN-Tracker as it achieves top performance on both tasks. Datasets will be released upon acceptance.
FS-MedSAM2: Exploring the Potential of SAM2 for Few-Shot Medical Image Segmentation without Fine-tuning
Sep 06, 2024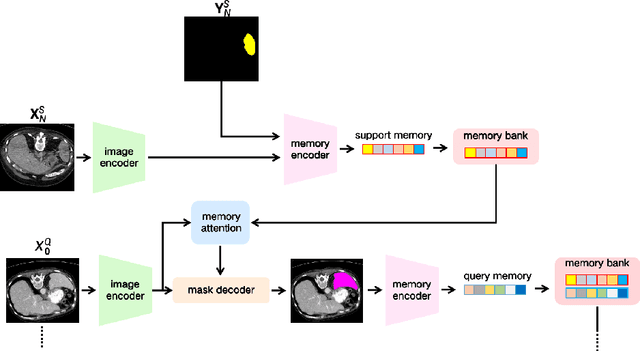

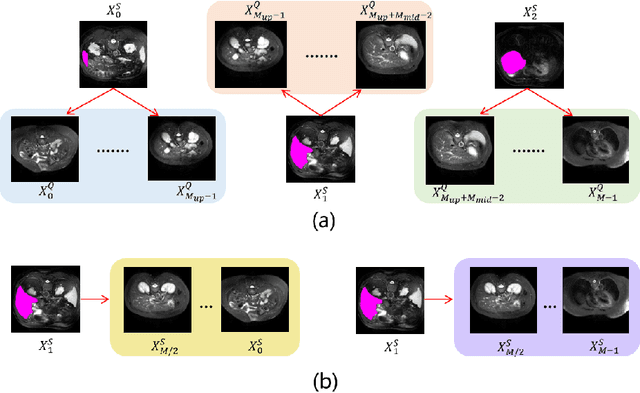

Abstract:The Segment Anything Model 2 (SAM2) has recently demonstrated exceptional performance in zero-shot prompt segmentation for natural images and videos. However, it faces significant challenges when applied to medical images. Since its release, many attempts have been made to adapt SAM2's segmentation capabilities to the medical imaging domain. These efforts typically involve using a substantial amount of labeled data to fine-tune the model's weights. In this paper, we explore SAM2 from a different perspective via making the full use of its trained memory attention module and its ability of processing mask prompts. We introduce FS-MedSAM2, a simple yet effective framework that enables SAM2 to achieve superior medical image segmentation in a few-shot setting, without the need for fine-tuning. Our framework outperforms the current state-of-the-arts on two publicly available medical image datasets. The code is available at https://github.com/DeepMed-Lab-ECNU/FS_MedSAM2.
Effective Lymph Nodes Detection in CT Scans Using Location Debiased Query Selection and Contrastive Query Representation in Transformer
Apr 04, 2024Abstract:Lymph node (LN) assessment is a critical, indispensable yet very challenging task in the routine clinical workflow of radiology and oncology. Accurate LN analysis is essential for cancer diagnosis, staging, and treatment planning. Finding scatteredly distributed, low-contrast clinically relevant LNs in 3D CT is difficult even for experienced physicians under high inter-observer variations. Previous automatic LN detection works typically yield limited recall and high false positives (FPs) due to adjacent anatomies with similar image intensities, shapes, or textures (vessels, muscles, esophagus, etc). In this work, we propose a new LN DEtection TRansformer, named LN-DETR, to achieve more accurate performance. By enhancing the 2D backbone with a multi-scale 2.5D feature fusion to incorporate 3D context explicitly, more importantly, we make two main contributions to improve the representation quality of LN queries. 1) Considering that LN boundaries are often unclear, an IoU prediction head and a location debiased query selection are proposed to select LN queries of higher localization accuracy as the decoder query's initialization. 2) To reduce FPs, query contrastive learning is employed to explicitly reinforce LN queries towards their best-matched ground-truth queries over unmatched query predictions. Trained and tested on 3D CT scans of 1067 patients (with 10,000+ labeled LNs) via combining seven LN datasets from different body parts (neck, chest, and abdomen) and pathologies/cancers, our method significantly improves the performance of previous leading methods by > 4-5% average recall at the same FP rates in both internal and external testing. We further evaluate on the universal lesion detection task using NIH DeepLesion benchmark, and our method achieves the top performance of 88.46% averaged recall across 0.5 to 4 FPs per image, compared with other leading reported results.
ReSynthDetect: A Fundus Anomaly Detection Network with Reconstruction and Synthetic Features
Dec 27, 2023Abstract:Detecting anomalies in fundus images through unsupervised methods is a challenging task due to the similarity between normal and abnormal tissues, as well as their indistinct boundaries. The current methods have limitations in accurately detecting subtle anomalies while avoiding false positives. To address these challenges, we propose the ReSynthDetect network which utilizes a reconstruction network for modeling normal images, and an anomaly generator that produces synthetic anomalies consistent with the appearance of fundus images. By combining the features of consistent anomaly generation and image reconstruction, our method is suited for detecting fundus abnormalities. The proposed approach has been extensively tested on benchmark datasets such as EyeQ and IDRiD, demonstrating state-of-the-art performance in both image-level and pixel-level anomaly detection. Our experiments indicate a substantial 9% improvement in AUROC on EyeQ and a significant 17.1% improvement in AUPR on IDRiD.
Source-Free Domain Adaptation for Medical Image Segmentation via Prototype-Anchored Feature Alignment and Contrastive Learning
Jul 19, 2023Abstract:Unsupervised domain adaptation (UDA) has increasingly gained interests for its capacity to transfer the knowledge learned from a labeled source domain to an unlabeled target domain. However, typical UDA methods require concurrent access to both the source and target domain data, which largely limits its application in medical scenarios where source data is often unavailable due to privacy concern. To tackle the source data-absent problem, we present a novel two-stage source-free domain adaptation (SFDA) framework for medical image segmentation, where only a well-trained source segmentation model and unlabeled target data are available during domain adaptation. Specifically, in the prototype-anchored feature alignment stage, we first utilize the weights of the pre-trained pixel-wise classifier as source prototypes, which preserve the information of source features. Then, we introduce the bi-directional transport to align the target features with class prototypes by minimizing its expected cost. On top of that, a contrastive learning stage is further devised to utilize those pixels with unreliable predictions for a more compact target feature distribution. Extensive experiments on a cross-modality medical segmentation task demonstrate the superiority of our method in large domain discrepancy settings compared with the state-of-the-art SFDA approaches and even some UDA methods. Code is available at https://github.com/CSCYQJ/MICCAI23-ProtoContra-SFDA.
Region and Spatial Aware Anomaly Detection for Fundus Images
Mar 07, 2023Abstract:Recently anomaly detection has drawn much attention in diagnosing ocular diseases. Most existing anomaly detection research in fundus images has relatively large anomaly scores in the salient retinal structures, such as blood vessels, optical cups and discs. In this paper, we propose a Region and Spatial Aware Anomaly Detection (ReSAD) method for fundus images, which obtains local region and long-range spatial information to reduce the false positives in the normal structure. ReSAD transfers a pre-trained model to extract the features of normal fundus images and applies the Region-and-Spatial-Aware feature Combination module (ReSC) for pixel-level features to build a memory bank. In the testing phase, ReSAD uses the memory bank to determine out-of-distribution samples as abnormalities. Our method significantly outperforms the existing anomaly detection methods for fundus images on two publicly benchmark datasets.
Coarse Retinal Lesion Annotations Refinement via Prototypical Learning
Aug 30, 2022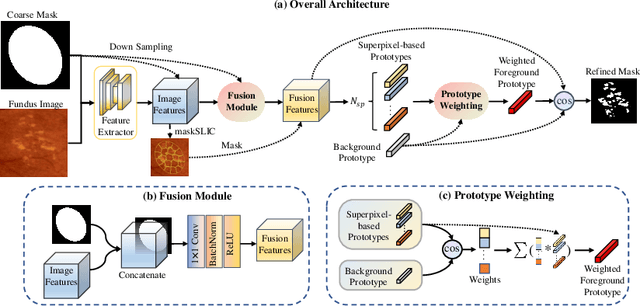
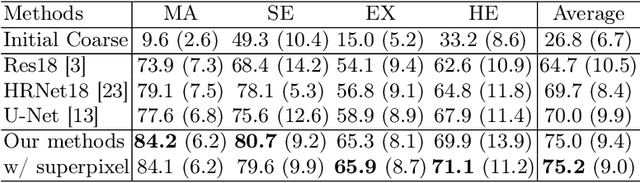
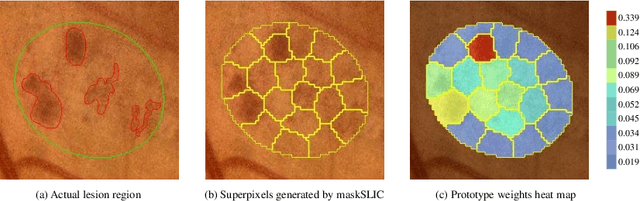
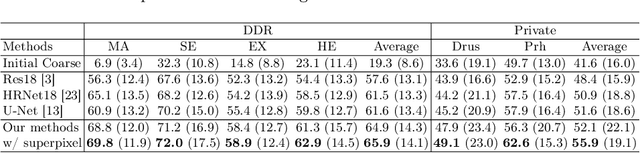
Abstract:Deep-learning-based approaches for retinal lesion segmentation often require an abundant amount of precise pixel-wise annotated data. However, coarse annotations such as circles or ellipses for outlining the lesion area can be six times more efficient than pixel-level annotation. Therefore, this paper proposes an annotation refinement network to convert a coarse annotation into a pixel-level segmentation mask. Our main novelty is the application of the prototype learning paradigm to enhance the generalization ability across different datasets or types of lesions. We also introduce a prototype weighing module to handle challenging cases where the lesion is overly small. The proposed method was trained on the publicly available IDRiD dataset and then generalized to the public DDR and our real-world private datasets. Experiments show that our approach substantially improved the initial coarse mask and outperformed the non-prototypical baseline by a large margin. Moreover, we demonstrate the usefulness of the prototype weighing module in both cross-dataset and cross-class settings.
GAMMA Challenge:Glaucoma grAding from Multi-Modality imAges
Feb 16, 2022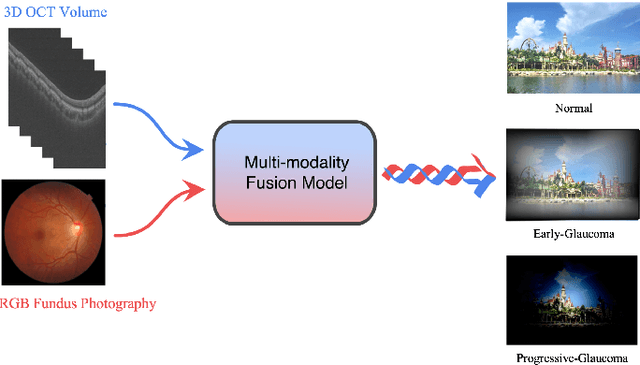
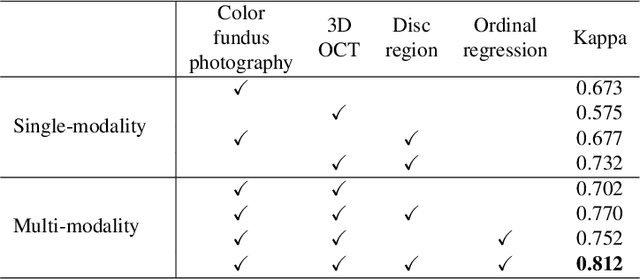
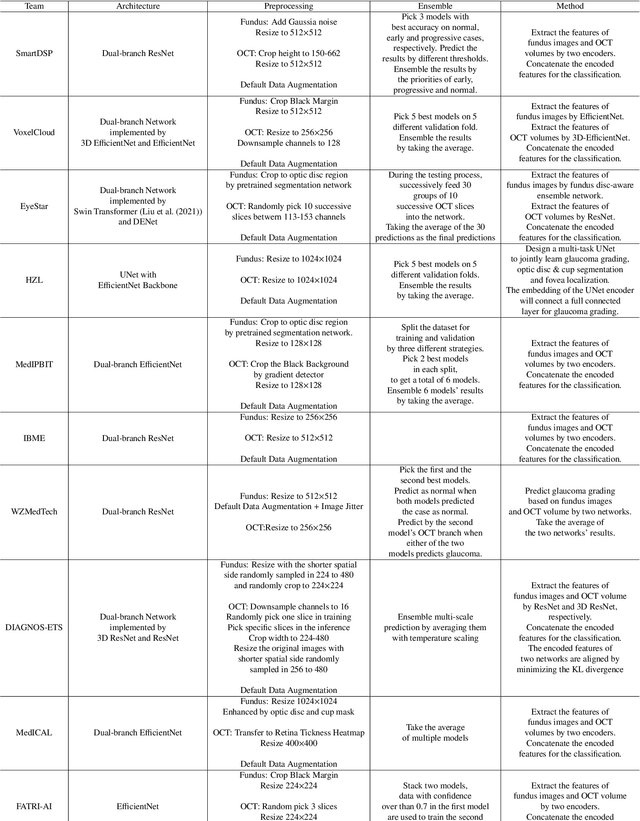
Abstract:Color fundus photography and Optical Coherence Tomography (OCT) are the two most cost-effective tools for glaucoma screening. Both two modalities of images have prominent biomarkers to indicate glaucoma suspected. Clinically, it is often recommended to take both of the screenings for a more accurate and reliable diagnosis. However, although numerous algorithms are proposed based on fundus images or OCT volumes in computer-aided diagnosis, there are still few methods leveraging both of the modalities for the glaucoma assessment. Inspired by the success of Retinal Fundus Glaucoma Challenge (REFUGE) we held previously, we set up the Glaucoma grAding from Multi-Modality imAges (GAMMA) Challenge to encourage the development of fundus \& OCT-based glaucoma grading. The primary task of the challenge is to grade glaucoma from both the 2D fundus images and 3D OCT scanning volumes. As part of GAMMA, we have publicly released a glaucoma annotated dataset with both 2D fundus color photography and 3D OCT volumes, which is the first multi-modality dataset for glaucoma grading. In addition, an evaluation framework is also established to evaluate the performance of the submitted methods. During the challenge, 1272 results were submitted, and finally, top-10 teams were selected to the final stage. We analysis their results and summarize their methods in the paper. Since all these teams submitted their source code in the challenge, a detailed ablation study is also conducted to verify the effectiveness of the particular modules proposed. We find many of the proposed techniques are practical for the clinical diagnosis of glaucoma. As the first in-depth study of fundus \& OCT multi-modality glaucoma grading, we believe the GAMMA Challenge will be an essential starting point for future research.
Bilateral-ViT for Robust Fovea Localization
Oct 19, 2021



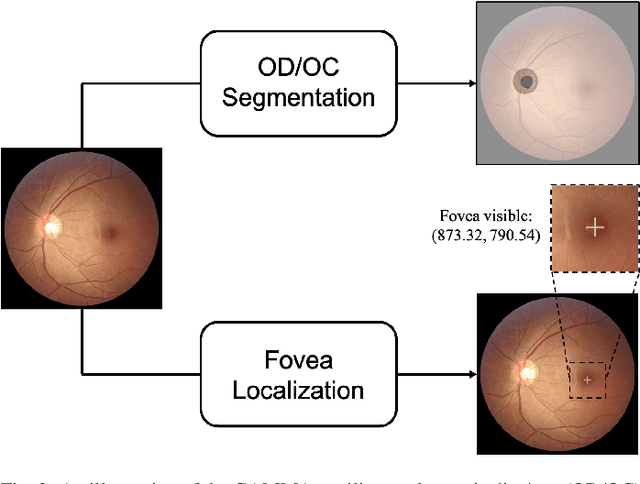
Abstract:The fovea is an important anatomical landmark of the retina. Detecting the location of the fovea is essential for the analysis of many retinal diseases. However, robust fovea localization remains a challenging problem, as the fovea region often appears fuzzy, and retina diseases may further obscure its appearance. This paper proposes a novel vision transformer (ViT) approach that integrates information both inside and outside the fovea region to achieve robust fovea localization. Our proposed network named Bilateral-Vision-Transformer (Bilateral-ViT) consists of two network branches: a transformer-based main network branch for integrating global context across the entire fundus image and a vessel branch for explicitly incorporating the structure of blood vessels. The encoded features from both network branches are subsequently merged with a customized multi-scale feature fusion (MFF) module. Our comprehensive experiments demonstrate that the proposed approach is significantly more robust for diseased images and establishes the new state of the arts on both Messidor and PALM datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge