Jingqi Niu
ReSynthDetect: A Fundus Anomaly Detection Network with Reconstruction and Synthetic Features
Dec 27, 2023Abstract:Detecting anomalies in fundus images through unsupervised methods is a challenging task due to the similarity between normal and abnormal tissues, as well as their indistinct boundaries. The current methods have limitations in accurately detecting subtle anomalies while avoiding false positives. To address these challenges, we propose the ReSynthDetect network which utilizes a reconstruction network for modeling normal images, and an anomaly generator that produces synthetic anomalies consistent with the appearance of fundus images. By combining the features of consistent anomaly generation and image reconstruction, our method is suited for detecting fundus abnormalities. The proposed approach has been extensively tested on benchmark datasets such as EyeQ and IDRiD, demonstrating state-of-the-art performance in both image-level and pixel-level anomaly detection. Our experiments indicate a substantial 9% improvement in AUROC on EyeQ and a significant 17.1% improvement in AUPR on IDRiD.
Region and Spatial Aware Anomaly Detection for Fundus Images
Mar 07, 2023Abstract:Recently anomaly detection has drawn much attention in diagnosing ocular diseases. Most existing anomaly detection research in fundus images has relatively large anomaly scores in the salient retinal structures, such as blood vessels, optical cups and discs. In this paper, we propose a Region and Spatial Aware Anomaly Detection (ReSAD) method for fundus images, which obtains local region and long-range spatial information to reduce the false positives in the normal structure. ReSAD transfers a pre-trained model to extract the features of normal fundus images and applies the Region-and-Spatial-Aware feature Combination module (ReSC) for pixel-level features to build a memory bank. In the testing phase, ReSAD uses the memory bank to determine out-of-distribution samples as abnormalities. Our method significantly outperforms the existing anomaly detection methods for fundus images on two publicly benchmark datasets.
Exploring Vanilla U-Net for Lesion Segmentation from Whole-body FDG-PET/CT Scans
Oct 14, 2022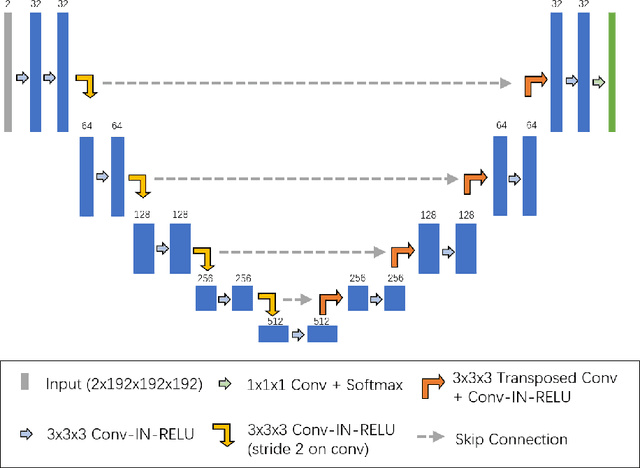
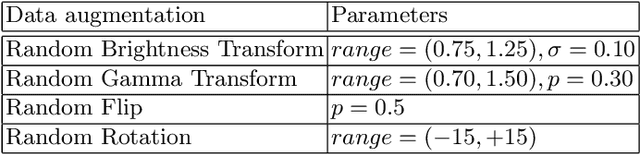
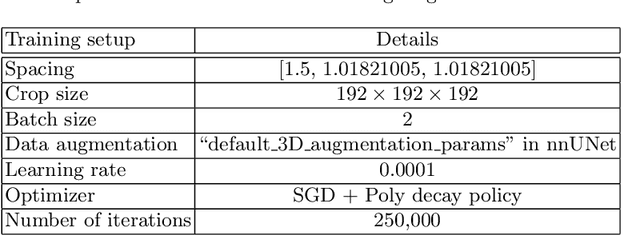
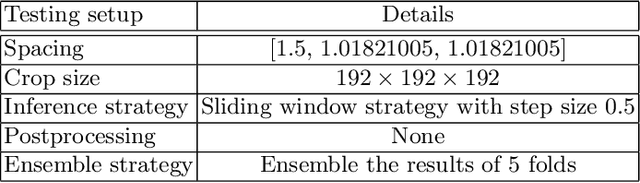
Abstract:Tumor lesion segmentation is one of the most important tasks in medical image analysis. In clinical practice, Fluorodeoxyglucose Positron-Emission Tomography~(FDG-PET) is a widely used technique to identify and quantify metabolically active tumors. However, since FDG-PET scans only provide metabolic information, healthy tissue or benign disease with irregular glucose consumption may be mistaken for cancer. To handle this challenge, PET is commonly combined with Computed Tomography~(CT), with the CT used to obtain the anatomic structure of the patient. The combination of PET-based metabolic and CT-based anatomic information can contribute to better tumor segmentation results. %Computed tomography~(CT) is a popular modality to illustrate the anatomic structure of the patient. The combination of PET and CT is promising to handle this challenge by utilizing metabolic and anatomic information. In this paper, we explore the potential of U-Net for lesion segmentation in whole-body FDG-PET/CT scans from three aspects, including network architecture, data preprocessing, and data augmentation. The experimental results demonstrate that the vanilla U-Net with proper input shape can achieve satisfactory performance. Specifically, our method achieves first place in both preliminary and final leaderboards of the autoPET 2022 challenge. Our code is available at https://github.com/Yejin0111/autoPET2022_Blackbean.
An evaluation of U-Net in Renal Structure Segmentation
Sep 06, 2022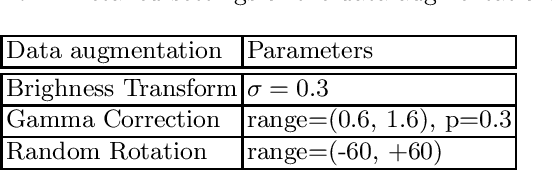
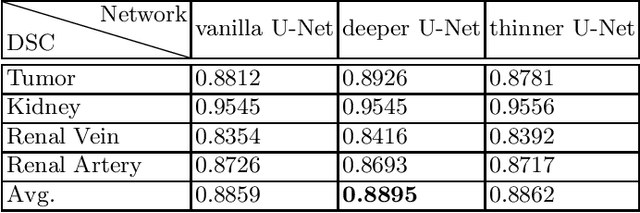
Abstract:Renal structure segmentation from computed tomography angiography~(CTA) is essential for many computer-assisted renal cancer treatment applications. Kidney PArsing~(KiPA 2022) Challenge aims to build a fine-grained multi-structure dataset and improve the segmentation of multiple renal structures. Recently, U-Net has dominated the medical image segmentation. In the KiPA challenge, we evaluated several U-Net variants and selected the best models for the final submission.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge