Kang Dang
EndoFlow-SLAM: Real-Time Endoscopic SLAM with Flow-Constrained Gaussian Splatting
Jun 26, 2025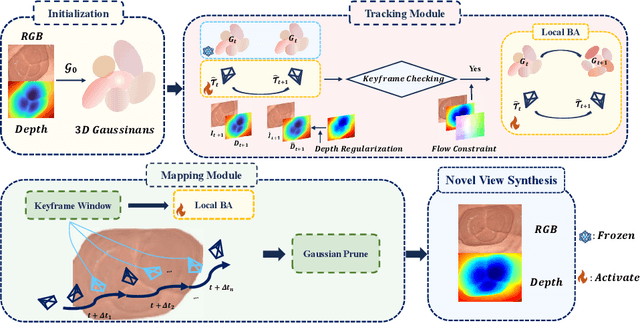
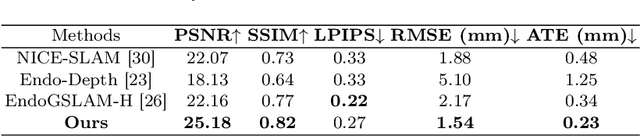
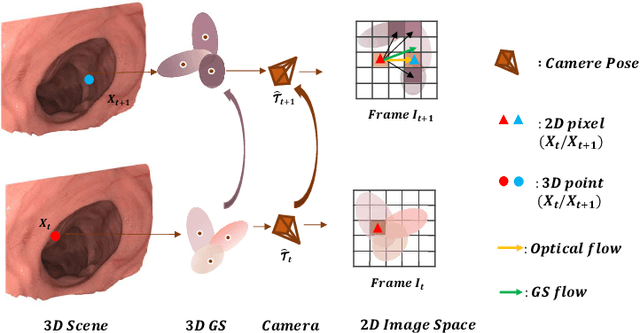
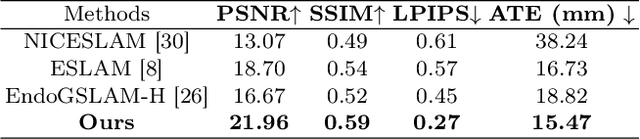
Abstract:Efficient three-dimensional reconstruction and real-time visualization are critical in surgical scenarios such as endoscopy. In recent years, 3D Gaussian Splatting (3DGS) has demonstrated remarkable performance in efficient 3D reconstruction and rendering. Most 3DGS-based Simultaneous Localization and Mapping (SLAM) methods only rely on the appearance constraints for optimizing both 3DGS and camera poses. However, in endoscopic scenarios, the challenges include photometric inconsistencies caused by non-Lambertian surfaces and dynamic motion from breathing affects the performance of SLAM systems. To address these issues, we additionally introduce optical flow loss as a geometric constraint, which effectively constrains both the 3D structure of the scene and the camera motion. Furthermore, we propose a depth regularisation strategy to mitigate the problem of photometric inconsistencies and ensure the validity of 3DGS depth rendering in endoscopic scenes. In addition, to improve scene representation in the SLAM system, we improve the 3DGS refinement strategy by focusing on viewpoints corresponding to Keyframes with suboptimal rendering quality frames, achieving better rendering results. Extensive experiments on the C3VD static dataset and the StereoMIS dynamic dataset demonstrate that our method outperforms existing state-of-the-art methods in novel view synthesis and pose estimation, exhibiting high performance in both static and dynamic surgical scenes. The source code will be publicly available upon paper acceptance.
MSWAL: 3D Multi-class Segmentation of Whole Abdominal Lesions Dataset
Mar 17, 2025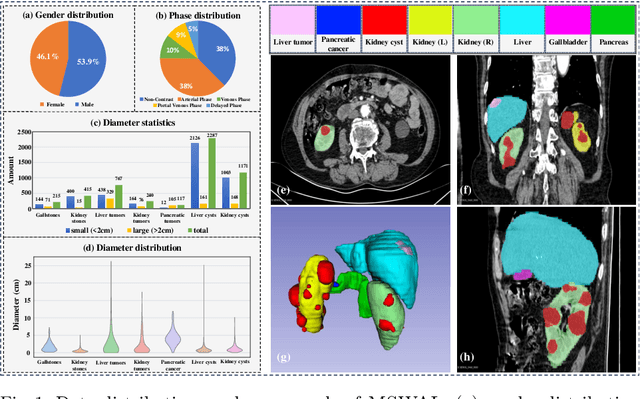
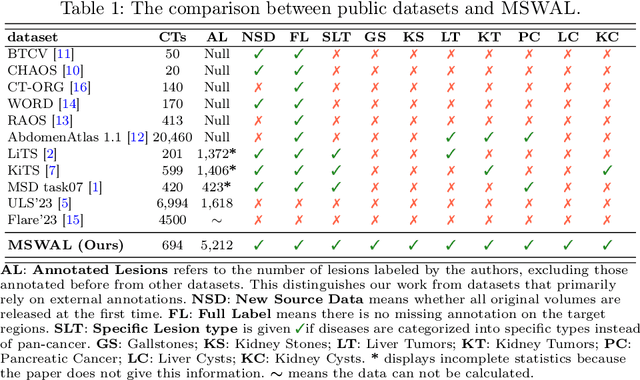
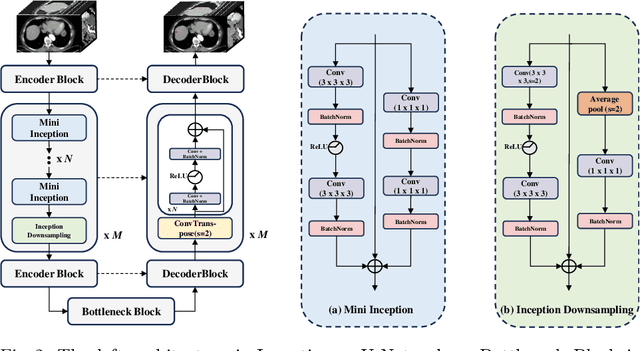
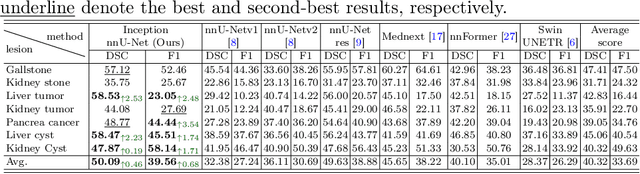
Abstract:With the significantly increasing incidence and prevalence of abdominal diseases, there is a need to embrace greater use of new innovations and technology for the diagnosis and treatment of patients. Although deep-learning methods have notably been developed to assist radiologists in diagnosing abdominal diseases, existing models have the restricted ability to segment common lesions in the abdomen due to missing annotations for typical abdominal pathologies in their training datasets. To address the limitation, we introduce MSWAL, the first 3D Multi-class Segmentation of the Whole Abdominal Lesions dataset, which broadens the coverage of various common lesion types, such as gallstones, kidney stones, liver tumors, kidney tumors, pancreatic cancer, liver cysts, and kidney cysts. With CT scans collected from 694 patients (191,417 slices) of different genders across various scanning phases, MSWAL demonstrates strong robustness and generalizability. The transfer learning experiment from MSWAL to two public datasets, LiTS and KiTS, effectively demonstrates consistent improvements, with Dice Similarity Coefficient (DSC) increase of 3.00% for liver tumors and 0.89% for kidney tumors, demonstrating that the comprehensive annotations and diverse lesion types in MSWAL facilitate effective learning across different domains and data distributions. Furthermore, we propose Inception nnU-Net, a novel segmentation framework that effectively integrates an Inception module with the nnU-Net architecture to extract information from different receptive fields, achieving significant enhancement in both voxel-level DSC and region-level F1 compared to the cutting-edge public algorithms on MSWAL. Our dataset will be released after being accepted, and the code is publicly released at https://github.com/tiuxuxsh76075/MSWAL-.
Beyond Words: AuralLLM and SignMST-C for Precise Sign Language Production and Bidirectional Accessibility
Jan 01, 2025Abstract:Although sign language recognition aids non-hearing-impaired understanding, many hearing-impaired individuals still rely on sign language alone due to limited literacy, underscoring the need for advanced sign language production and translation (SLP and SLT) systems. In the field of sign language production, the lack of adequate models and datasets restricts practical applications. Existing models face challenges in production accuracy and pose control, making it difficult to provide fluent sign language expressions across diverse scenarios. Additionally, data resources are scarce, particularly high-quality datasets with complete sign vocabulary and pose annotations. To address these issues, we introduce CNText2Sign and CNSign, comprehensive datasets to benchmark SLP and SLT, respectively, with CNText2Sign covering gloss and landmark mappings for SLP, and CNSign providing extensive video-to-text data for SLT. To improve the accuracy and applicability of sign language systems, we propose the AuraLLM and SignMST-C models. AuraLLM, incorporating LoRA and RAG techniques, achieves a BLEU-4 score of 50.41 on the CNText2Sign dataset, enabling precise control over gesture semantics and motion. SignMST-C employs self-supervised rapid motion video pretraining, achieving a BLEU-4 score of 31.03/32.08 on the PHOENIX2014-T benchmark, setting a new state-of-the-art. These models establish robust baselines for the datasets released for their respective tasks.
Decoding the Flow: CauseMotion for Emotional Causality Analysis in Long-form Conversations
Jan 01, 2025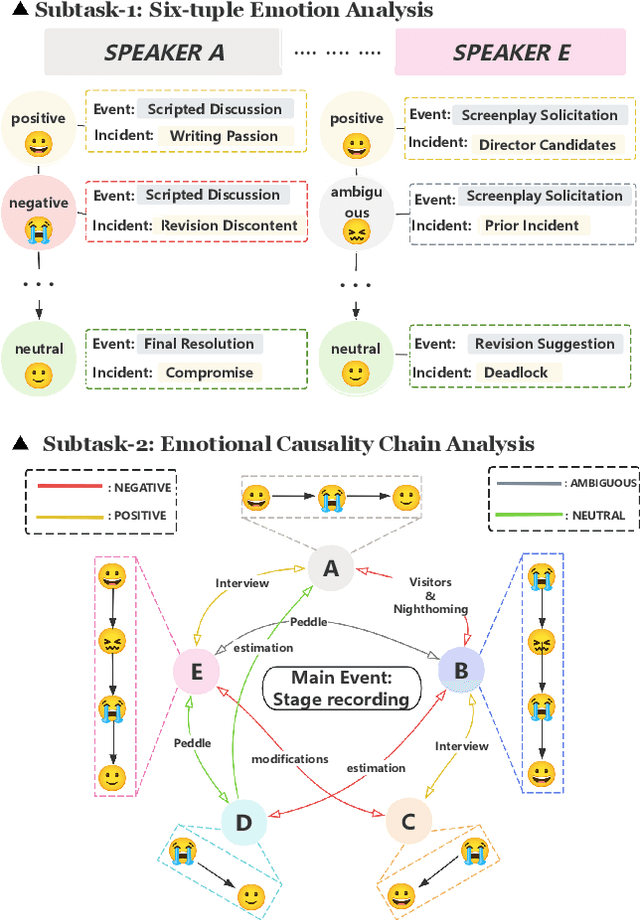
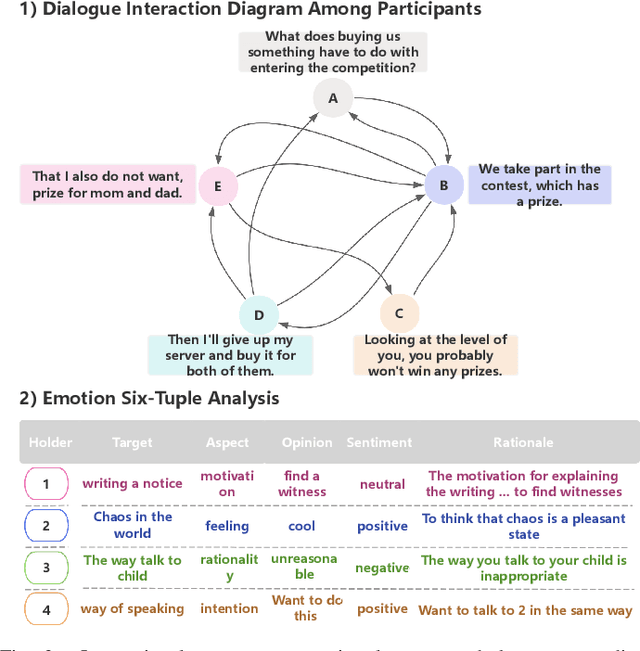
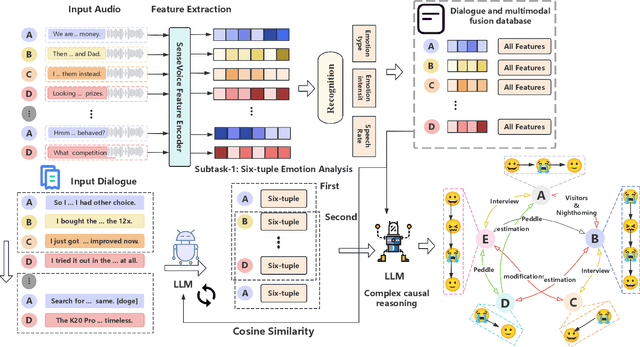
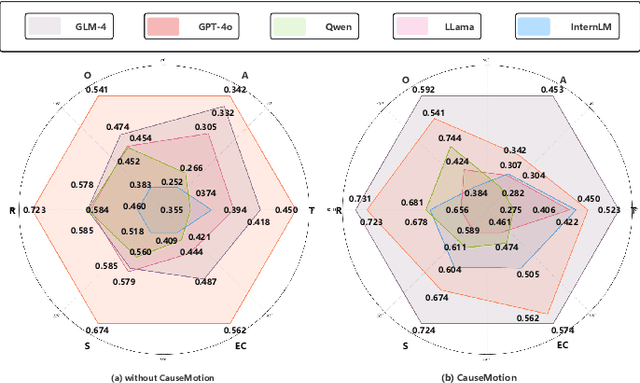
Abstract:Long-sequence causal reasoning seeks to uncover causal relationships within extended time series data but is hindered by complex dependencies and the challenges of validating causal links. To address the limitations of large-scale language models (e.g., GPT-4) in capturing intricate emotional causality within extended dialogues, we propose CauseMotion, a long-sequence emotional causal reasoning framework grounded in Retrieval-Augmented Generation (RAG) and multimodal fusion. Unlike conventional methods relying only on textual information, CauseMotion enriches semantic representations by incorporating audio-derived features-vocal emotion, emotional intensity, and speech rate-into textual modalities. By integrating RAG with a sliding window mechanism, it effectively retrieves and leverages contextually relevant dialogue segments, thus enabling the inference of complex emotional causal chains spanning multiple conversational turns. To evaluate its effectiveness, we constructed the first benchmark dataset dedicated to long-sequence emotional causal reasoning, featuring dialogues with over 70 turns. Experimental results demonstrate that the proposed RAG-based multimodal integrated approach, the efficacy of substantially enhances both the depth of emotional understanding and the causal inference capabilities of large-scale language models. A GLM-4 integrated with CauseMotion achieves an 8.7% improvement in causal accuracy over the original model and surpasses GPT-4o by 1.2%. Additionally, on the publicly available DiaASQ dataset, CauseMotion-GLM-4 achieves state-of-the-art results in accuracy, F1 score, and causal reasoning accuracy.
Modality-Aware Shot Relating and Comparing for Video Scene Detection
Dec 23, 2024



Abstract:Video scene detection involves assessing whether each shot and its surroundings belong to the same scene. Achieving this requires meticulously correlating multi-modal cues, $\it{e.g.}$ visual entity and place modalities, among shots and comparing semantic changes around each shot. However, most methods treat multi-modal semantics equally and do not examine contextual differences between the two sides of a shot, leading to sub-optimal detection performance. In this paper, we propose the $\bf{M}$odality-$\bf{A}$ware $\bf{S}$hot $\bf{R}$elating and $\bf{C}$omparing approach (MASRC), which enables relating shots per their own characteristics of visual entity and place modalities, as well as comparing multi-shots similarities to have scene changes explicitly encoded. Specifically, to fully harness the potential of visual entity and place modalities in modeling shot relations, we mine long-term shot correlations from entity semantics while simultaneously revealing short-term shot correlations from place semantics. In this way, we can learn distinctive shot features that consolidate coherence within scenes and amplify distinguishability across scenes. Once equipped with distinctive shot features, we further encode the relations between preceding and succeeding shots of each target shot by similarity convolution, aiding in the identification of scene ending shots. We validate the broad applicability of the proposed components in MASRC. Extensive experimental results on public benchmark datasets demonstrate that the proposed MASRC significantly advances video scene detection.
ReSynthDetect: A Fundus Anomaly Detection Network with Reconstruction and Synthetic Features
Dec 27, 2023Abstract:Detecting anomalies in fundus images through unsupervised methods is a challenging task due to the similarity between normal and abnormal tissues, as well as their indistinct boundaries. The current methods have limitations in accurately detecting subtle anomalies while avoiding false positives. To address these challenges, we propose the ReSynthDetect network which utilizes a reconstruction network for modeling normal images, and an anomaly generator that produces synthetic anomalies consistent with the appearance of fundus images. By combining the features of consistent anomaly generation and image reconstruction, our method is suited for detecting fundus abnormalities. The proposed approach has been extensively tested on benchmark datasets such as EyeQ and IDRiD, demonstrating state-of-the-art performance in both image-level and pixel-level anomaly detection. Our experiments indicate a substantial 9% improvement in AUROC on EyeQ and a significant 17.1% improvement in AUPR on IDRiD.
Source-Free Domain Adaptation for Medical Image Segmentation via Prototype-Anchored Feature Alignment and Contrastive Learning
Jul 19, 2023Abstract:Unsupervised domain adaptation (UDA) has increasingly gained interests for its capacity to transfer the knowledge learned from a labeled source domain to an unlabeled target domain. However, typical UDA methods require concurrent access to both the source and target domain data, which largely limits its application in medical scenarios where source data is often unavailable due to privacy concern. To tackle the source data-absent problem, we present a novel two-stage source-free domain adaptation (SFDA) framework for medical image segmentation, where only a well-trained source segmentation model and unlabeled target data are available during domain adaptation. Specifically, in the prototype-anchored feature alignment stage, we first utilize the weights of the pre-trained pixel-wise classifier as source prototypes, which preserve the information of source features. Then, we introduce the bi-directional transport to align the target features with class prototypes by minimizing its expected cost. On top of that, a contrastive learning stage is further devised to utilize those pixels with unreliable predictions for a more compact target feature distribution. Extensive experiments on a cross-modality medical segmentation task demonstrate the superiority of our method in large domain discrepancy settings compared with the state-of-the-art SFDA approaches and even some UDA methods. Code is available at https://github.com/CSCYQJ/MICCAI23-ProtoContra-SFDA.
Region and Spatial Aware Anomaly Detection for Fundus Images
Mar 07, 2023Abstract:Recently anomaly detection has drawn much attention in diagnosing ocular diseases. Most existing anomaly detection research in fundus images has relatively large anomaly scores in the salient retinal structures, such as blood vessels, optical cups and discs. In this paper, we propose a Region and Spatial Aware Anomaly Detection (ReSAD) method for fundus images, which obtains local region and long-range spatial information to reduce the false positives in the normal structure. ReSAD transfers a pre-trained model to extract the features of normal fundus images and applies the Region-and-Spatial-Aware feature Combination module (ReSC) for pixel-level features to build a memory bank. In the testing phase, ReSAD uses the memory bank to determine out-of-distribution samples as abnormalities. Our method significantly outperforms the existing anomaly detection methods for fundus images on two publicly benchmark datasets.
Bilateral-Fuser: A Novel Multi-cue Fusion Architecture with Anatomical-aware Tokens for Fovea Localization
Feb 14, 2023



Abstract:Accurate localization of fovea is one of the primary steps in analyzing retinal diseases since it helps prevent irreversible vision loss. Although current deep learning-based methods achieve better performance than traditional methods, there still remain challenges such as utilizing anatomical landmarks insufficiently, sensitivity to diseased retinal images and various image conditions. In this paper, we propose a novel transformer-based architecture (Bilateral-Fuser) for multi-cue fusion. This architecture explicitly incorporates long-range connections and global features using retina and vessel distributions for robust fovea localization. We introduce a spatial attention mechanism in the dual-stream encoder for extracting and fusing self-learned anatomical information. This design focuses more on features distributed along blood vessels and significantly decreases computational costs by reducing token numbers. Our comprehensive experiments show that the proposed architecture achieves state-of-the-art performance on two public and one large-scale private datasets. We also present that the Bilateral-Fuser is more robust on both normal and diseased retina images and has better generalization capacity in cross-dataset experiments.
Coarse Retinal Lesion Annotations Refinement via Prototypical Learning
Aug 30, 2022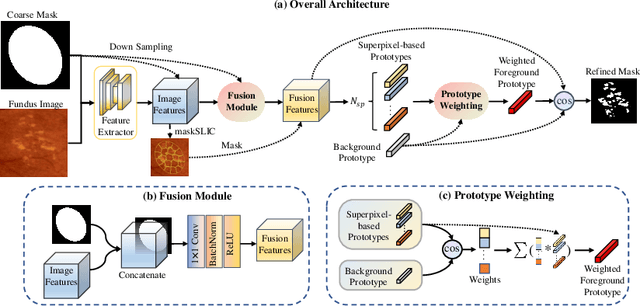
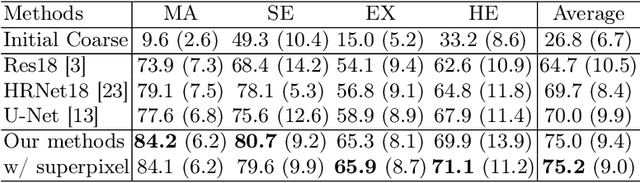
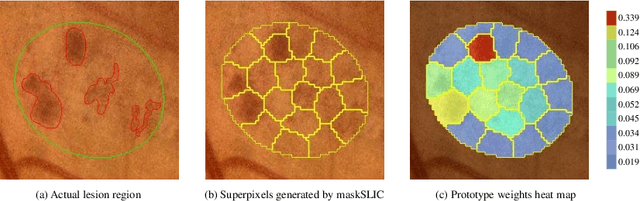
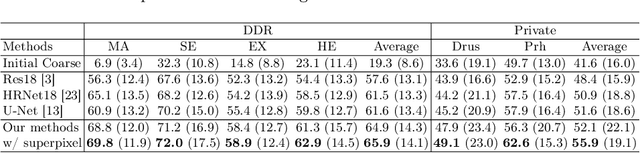
Abstract:Deep-learning-based approaches for retinal lesion segmentation often require an abundant amount of precise pixel-wise annotated data. However, coarse annotations such as circles or ellipses for outlining the lesion area can be six times more efficient than pixel-level annotation. Therefore, this paper proposes an annotation refinement network to convert a coarse annotation into a pixel-level segmentation mask. Our main novelty is the application of the prototype learning paradigm to enhance the generalization ability across different datasets or types of lesions. We also introduce a prototype weighing module to handle challenging cases where the lesion is overly small. The proposed method was trained on the publicly available IDRiD dataset and then generalized to the public DDR and our real-world private datasets. Experiments show that our approach substantially improved the initial coarse mask and outperformed the non-prototypical baseline by a large margin. Moreover, we demonstrate the usefulness of the prototype weighing module in both cross-dataset and cross-class settings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge