Xiaowei Ding
Lamps: Learning Anatomy from Multiple Perspectives via Self-supervision in Chest Radiographs
Jan 02, 2026Abstract:Foundation models have been successful in natural language processing and computer vision because they are capable of capturing the underlying structures (foundation) of natural languages. However, in medical imaging, the key foundation lies in human anatomy, as these images directly represent the internal structures of the body, reflecting the consistency, coherence, and hierarchy of human anatomy. Yet, existing self-supervised learning (SSL) methods often overlook these perspectives, limiting their ability to effectively learn anatomical features. To overcome the limitation, we built Lamps (learning anatomy from multiple perspectives via self-supervision) pre-trained on large-scale chest radiographs by harmoniously utilizing the consistency, coherence, and hierarchy of human anatomy as the supervision signal. Extensive experiments across 10 datasets evaluated through fine-tuning and emergent property analysis demonstrate Lamps' superior robustness, transferability, and clinical potential when compared to 10 baseline models. By learning from multiple perspectives, Lamps presents a unique opportunity for foundation models to develop meaningful, robust representations that are aligned with the structure of human anatomy.
Learning Anatomy from Multiple Perspectives via Self-supervision in Chest Radiographs
Dec 28, 2025Abstract:Foundation models have been successful in natural language processing and computer vision because they are capable of capturing the underlying structures (foundation) of natural languages. However, in medical imaging, the key foundation lies in human anatomy, as these images directly represent the internal structures of the body, reflecting the consistency, coherence, and hierarchy of human anatomy. Yet, existing self-supervised learning (SSL) methods often overlook these perspectives, limiting their ability to effectively learn anatomical features. To overcome the limitation, we built Lamps (learning anatomy from multiple perspectives via self-supervision) pre-trained on large-scale chest radiographs by harmoniously utilizing the consistency, coherence, and hierarchy of human anatomy as the supervision signal. Extensive experiments across 10 datasets evaluated through fine-tuning and emergent property analysis demonstrate Lamps' superior robustness, transferability, and clinical potential when compared to 10 baseline models. By learning from multiple perspectives, Lamps presents a unique opportunity for foundation models to develop meaningful, robust representations that are aligned with the structure of human anatomy.
From Slices to Sequences: Autoregressive Tracking Transformer for Cohesive and Consistent 3D Lymph Node Detection in CT Scans
Mar 11, 2025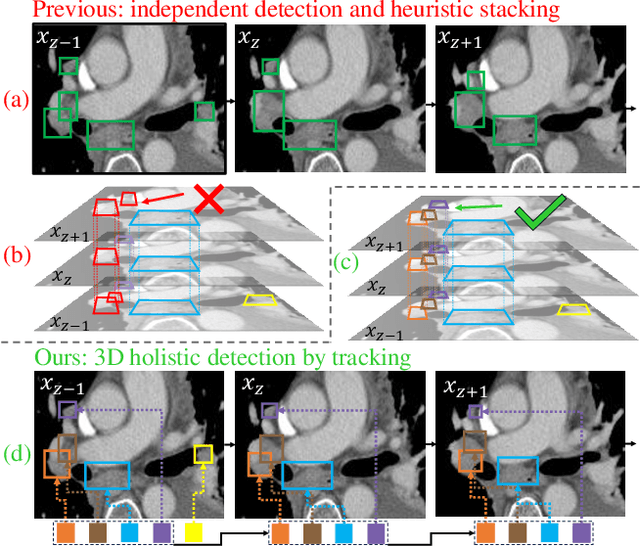

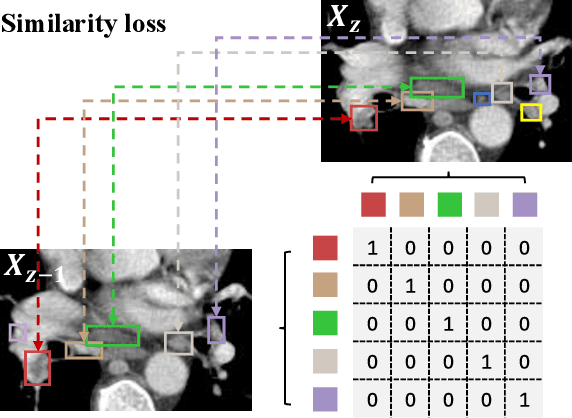
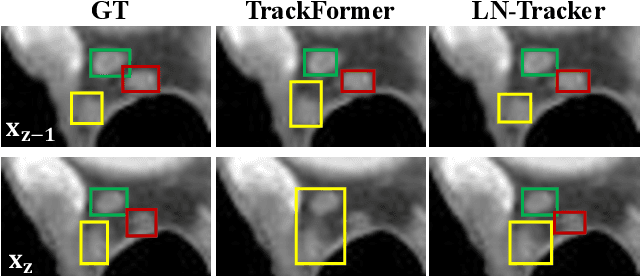
Abstract:Lymph node (LN) assessment is an essential task in the routine radiology workflow, providing valuable insights for cancer staging, treatment planning and beyond. Identifying scatteredly-distributed and low-contrast LNs in 3D CT scans is highly challenging, even for experienced clinicians. Previous lesion and LN detection methods demonstrate effectiveness of 2.5D approaches (i.e, using 2D network with multi-slice inputs), leveraging pretrained 2D model weights and showing improved accuracy as compared to separate 2D or 3D detectors. However, slice-based 2.5D detectors do not explicitly model inter-slice consistency for LN as a 3D object, requiring heuristic post-merging steps to generate final 3D LN instances, which can involve tuning a set of parameters for each dataset. In this work, we formulate 3D LN detection as a tracking task and propose LN-Tracker, a novel LN tracking transformer, for joint end-to-end detection and 3D instance association. Built upon DETR-based detector, LN-Tracker decouples transformer decoder's query into the track and detection groups, where the track query autoregressively follows previously tracked LN instances along the z-axis of a CT scan. We design a new transformer decoder with masked attention module to align track query's content to the context of current slice, meanwhile preserving detection query's high accuracy in current slice. An inter-slice similarity loss is introduced to encourage cohesive LN association between slices. Extensive evaluation on four lymph node datasets shows LN-Tracker's superior performance, with at least 2.7% gain in average sensitivity when compared to other top 3D/2.5D detectors. Further validation on public lung nodule and prostate tumor detection tasks confirms the generalizability of LN-Tracker as it achieves top performance on both tasks. Datasets will be released upon acceptance.
ACE: Anatomically Consistent Embeddings in Composition and Decomposition
Jan 17, 2025Abstract:Medical images acquired from standardized protocols show consistent macroscopic or microscopic anatomical structures, and these structures consist of composable/decomposable organs and tissues, but existing self-supervised learning (SSL) methods do not appreciate such composable/decomposable structure attributes inherent to medical images. To overcome this limitation, this paper introduces a novel SSL approach called ACE to learn anatomically consistent embedding via composition and decomposition with two key branches: (1) global consistency, capturing discriminative macro-structures via extracting global features; (2) local consistency, learning fine-grained anatomical details from composable/decomposable patch features via corresponding matrix matching. Experimental results across 6 datasets 2 backbones, evaluated in few-shot learning, fine-tuning, and property analysis, show ACE's superior robustness, transferability, and clinical potential. The innovations of our ACE lie in grid-wise image cropping, leveraging the intrinsic properties of compositionality and decompositionality of medical images, bridging the semantic gap from high-level pathologies to low-level tissue anomalies, and providing a new SSL method for medical imaging.
Effective Lymph Nodes Detection in CT Scans Using Location Debiased Query Selection and Contrastive Query Representation in Transformer
Apr 04, 2024Abstract:Lymph node (LN) assessment is a critical, indispensable yet very challenging task in the routine clinical workflow of radiology and oncology. Accurate LN analysis is essential for cancer diagnosis, staging, and treatment planning. Finding scatteredly distributed, low-contrast clinically relevant LNs in 3D CT is difficult even for experienced physicians under high inter-observer variations. Previous automatic LN detection works typically yield limited recall and high false positives (FPs) due to adjacent anatomies with similar image intensities, shapes, or textures (vessels, muscles, esophagus, etc). In this work, we propose a new LN DEtection TRansformer, named LN-DETR, to achieve more accurate performance. By enhancing the 2D backbone with a multi-scale 2.5D feature fusion to incorporate 3D context explicitly, more importantly, we make two main contributions to improve the representation quality of LN queries. 1) Considering that LN boundaries are often unclear, an IoU prediction head and a location debiased query selection are proposed to select LN queries of higher localization accuracy as the decoder query's initialization. 2) To reduce FPs, query contrastive learning is employed to explicitly reinforce LN queries towards their best-matched ground-truth queries over unmatched query predictions. Trained and tested on 3D CT scans of 1067 patients (with 10,000+ labeled LNs) via combining seven LN datasets from different body parts (neck, chest, and abdomen) and pathologies/cancers, our method significantly improves the performance of previous leading methods by > 4-5% average recall at the same FP rates in both internal and external testing. We further evaluate on the universal lesion detection task using NIH DeepLesion benchmark, and our method achieves the top performance of 88.46% averaged recall across 0.5 to 4 FPs per image, compared with other leading reported results.
ReSynthDetect: A Fundus Anomaly Detection Network with Reconstruction and Synthetic Features
Dec 27, 2023Abstract:Detecting anomalies in fundus images through unsupervised methods is a challenging task due to the similarity between normal and abnormal tissues, as well as their indistinct boundaries. The current methods have limitations in accurately detecting subtle anomalies while avoiding false positives. To address these challenges, we propose the ReSynthDetect network which utilizes a reconstruction network for modeling normal images, and an anomaly generator that produces synthetic anomalies consistent with the appearance of fundus images. By combining the features of consistent anomaly generation and image reconstruction, our method is suited for detecting fundus abnormalities. The proposed approach has been extensively tested on benchmark datasets such as EyeQ and IDRiD, demonstrating state-of-the-art performance in both image-level and pixel-level anomaly detection. Our experiments indicate a substantial 9% improvement in AUROC on EyeQ and a significant 17.1% improvement in AUPR on IDRiD.
Learning Anatomically Consistent Embedding for Chest Radiography
Dec 01, 2023Abstract:Self-supervised learning (SSL) approaches have recently shown substantial success in learning visual representations from unannotated images. Compared with photographic images, medical images acquired with the same imaging protocol exhibit high consistency in anatomy. To exploit this anatomical consistency, this paper introduces a novel SSL approach, called PEAC (patch embedding of anatomical consistency), for medical image analysis. Specifically, in this paper, we propose to learn global and local consistencies via stable grid-based matching, transfer pre-trained PEAC models to diverse downstream tasks, and extensively demonstrate that (1) PEAC achieves significantly better performance than the existing state-of-the-art fully/self-supervised methods, and (2) PEAC captures the anatomical structure consistency across views of the same patient and across patients of different genders, weights, and healthy statuses, which enhances the interpretability of our method for medical image analysis.
Source-Free Domain Adaptation for Medical Image Segmentation via Prototype-Anchored Feature Alignment and Contrastive Learning
Jul 19, 2023Abstract:Unsupervised domain adaptation (UDA) has increasingly gained interests for its capacity to transfer the knowledge learned from a labeled source domain to an unlabeled target domain. However, typical UDA methods require concurrent access to both the source and target domain data, which largely limits its application in medical scenarios where source data is often unavailable due to privacy concern. To tackle the source data-absent problem, we present a novel two-stage source-free domain adaptation (SFDA) framework for medical image segmentation, where only a well-trained source segmentation model and unlabeled target data are available during domain adaptation. Specifically, in the prototype-anchored feature alignment stage, we first utilize the weights of the pre-trained pixel-wise classifier as source prototypes, which preserve the information of source features. Then, we introduce the bi-directional transport to align the target features with class prototypes by minimizing its expected cost. On top of that, a contrastive learning stage is further devised to utilize those pixels with unreliable predictions for a more compact target feature distribution. Extensive experiments on a cross-modality medical segmentation task demonstrate the superiority of our method in large domain discrepancy settings compared with the state-of-the-art SFDA approaches and even some UDA methods. Code is available at https://github.com/CSCYQJ/MICCAI23-ProtoContra-SFDA.
Region and Spatial Aware Anomaly Detection for Fundus Images
Mar 07, 2023Abstract:Recently anomaly detection has drawn much attention in diagnosing ocular diseases. Most existing anomaly detection research in fundus images has relatively large anomaly scores in the salient retinal structures, such as blood vessels, optical cups and discs. In this paper, we propose a Region and Spatial Aware Anomaly Detection (ReSAD) method for fundus images, which obtains local region and long-range spatial information to reduce the false positives in the normal structure. ReSAD transfers a pre-trained model to extract the features of normal fundus images and applies the Region-and-Spatial-Aware feature Combination module (ReSC) for pixel-level features to build a memory bank. In the testing phase, ReSAD uses the memory bank to determine out-of-distribution samples as abnormalities. Our method significantly outperforms the existing anomaly detection methods for fundus images on two publicly benchmark datasets.
Bilateral-Fuser: A Novel Multi-cue Fusion Architecture with Anatomical-aware Tokens for Fovea Localization
Feb 14, 2023



Abstract:Accurate localization of fovea is one of the primary steps in analyzing retinal diseases since it helps prevent irreversible vision loss. Although current deep learning-based methods achieve better performance than traditional methods, there still remain challenges such as utilizing anatomical landmarks insufficiently, sensitivity to diseased retinal images and various image conditions. In this paper, we propose a novel transformer-based architecture (Bilateral-Fuser) for multi-cue fusion. This architecture explicitly incorporates long-range connections and global features using retina and vessel distributions for robust fovea localization. We introduce a spatial attention mechanism in the dual-stream encoder for extracting and fusing self-learned anatomical information. This design focuses more on features distributed along blood vessels and significantly decreases computational costs by reducing token numbers. Our comprehensive experiments show that the proposed architecture achieves state-of-the-art performance on two public and one large-scale private datasets. We also present that the Bilateral-Fuser is more robust on both normal and diseased retina images and has better generalization capacity in cross-dataset experiments.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge