Dandan Zheng
TriC-Motion: Tri-Domain Causal Modeling Grounded Text-to-Motion Generation
Feb 09, 2026Abstract:Text-to-motion generation, a rapidly evolving field in computer vision, aims to produce realistic and text-aligned motion sequences. Current methods primarily focus on spatial-temporal modeling or independent frequency domain analysis, lacking a unified framework for joint optimization across spatial, temporal, and frequency domains. This limitation hinders the model's ability to leverage information from all domains simultaneously, leading to suboptimal generation quality. Additionally, in motion generation frameworks, motion-irrelevant cues caused by noise are often entangled with features that contribute positively to generation, thereby leading to motion distortion. To address these issues, we propose Tri-Domain Causal Text-to-Motion Generation (TriC-Motion), a novel diffusion-based framework integrating spatial-temporal-frequency-domain modeling with causal intervention. TriC-Motion includes three core modeling modules for domain-specific modeling, namely Temporal Motion Encoding, Spatial Topology Modeling, and Hybrid Frequency Analysis. After comprehensive modeling, a Score-guided Tri-domain Fusion module integrates valuable information from the triple domains, simultaneously ensuring temporal consistency, spatial topology, motion trends, and dynamics. Moreover, the Causality-based Counterfactual Motion Disentangler is meticulously designed to expose motion-irrelevant cues to eliminate noise, disentangling the real modeling contributions of each domain for superior generation. Extensive experimental results validate that TriC-Motion achieves superior performance compared to state-of-the-art methods, attaining an outstanding R@1 of 0.612 on the HumanML3D dataset. These results demonstrate its capability to generate high-fidelity, coherent, diverse, and text-aligned motion sequences. Code is available at: https://caoyiyang1105.github.io/TriC-Motion/.
GO-MLVTON: Garment Occlusion-Aware Multi-Layer Virtual Try-On with Diffusion Models
Jan 20, 2026Abstract:Existing Image-based virtual try-on (VTON) methods primarily focus on single-layer or multi-garment VTON, neglecting multi-layer VTON (ML-VTON), which involves dressing multiple layers of garments onto the human body with realistic deformation and layering to generate visually plausible outcomes. The main challenge lies in accurately modeling occlusion relationships between inner and outer garments to reduce interference from redundant inner garment features. To address this, we propose GO-MLVTON, the first multi-layer VTON method, introducing the Garment Occlusion Learning module to learn occlusion relationships and the StableDiffusion-based Garment Morphing & Fitting module to deform and fit garments onto the human body, producing high-quality multi-layer try-on results. Additionally, we present the MLG dataset for this task and propose a new metric named Layered Appearance Coherence Difference (LACD) for evaluation. Extensive experiments demonstrate the state-of-the-art performance of GO-MLVTON. Project page: https://upyuyang.github.io/go-mlvton/.
3SGen: Unified Subject, Style, and Structure-Driven Image Generation with Adaptive Task-specific Memory
Dec 22, 2025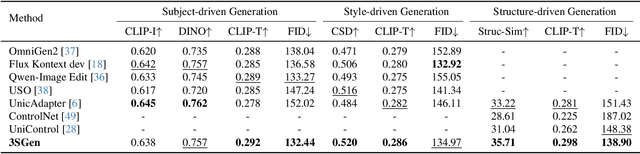
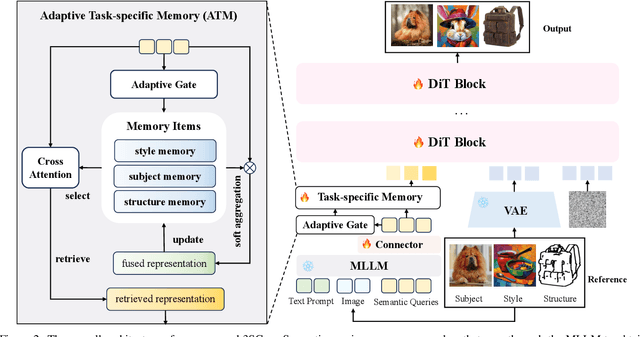
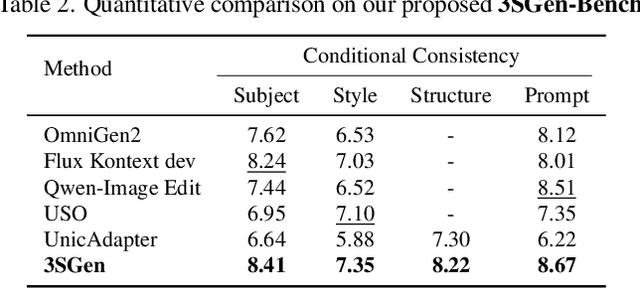
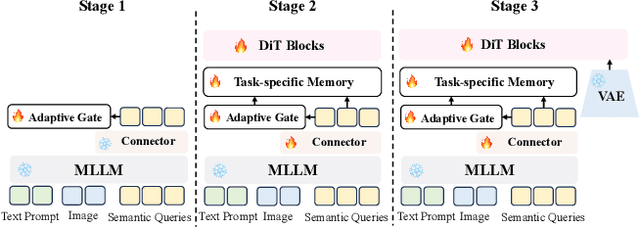
Abstract:Recent image generation approaches often address subject, style, and structure-driven conditioning in isolation, leading to feature entanglement and limited task transferability. In this paper, we introduce 3SGen, a task-aware unified framework that performs all three conditioning modes within a single model. 3SGen employs an MLLM equipped with learnable semantic queries to align text-image semantics, complemented by a VAE branch that preserves fine-grained visual details. At its core, an Adaptive Task-specific Memory (ATM) module dynamically disentangles, stores, and retrieves condition-specific priors, such as identity for subjects, textures for styles, and spatial layouts for structures, via a lightweight gating mechanism along with several scalable memory items. This design mitigates inter-task interference and naturally scales to compositional inputs. In addition, we propose 3SGen-Bench, a unified image-driven generation benchmark with standardized metrics for evaluating cross-task fidelity and controllability. Extensive experiments on our proposed 3SGen-Bench and other public benchmarks demonstrate our superior performance across diverse image-driven generation tasks.
ARGenSeg: Image Segmentation with Autoregressive Image Generation Model
Oct 23, 2025
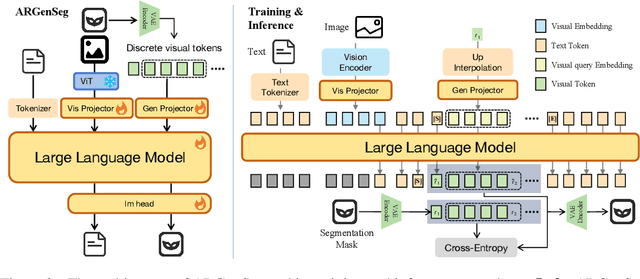
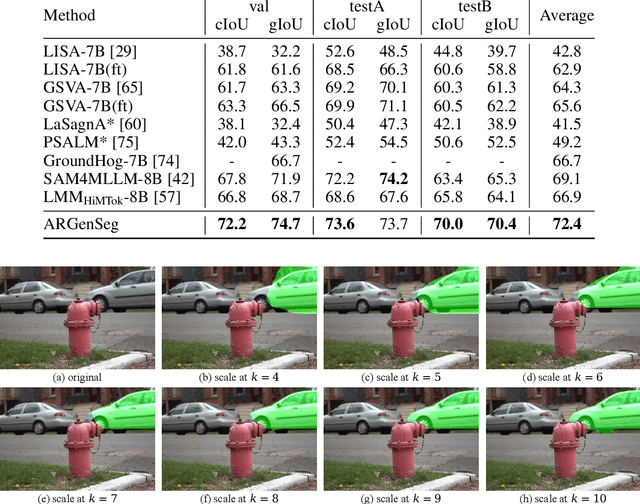

Abstract:We propose a novel AutoRegressive Generation-based paradigm for image Segmentation (ARGenSeg), achieving multimodal understanding and pixel-level perception within a unified framework. Prior works integrating image segmentation into multimodal large language models (MLLMs) typically employ either boundary points representation or dedicated segmentation heads. These methods rely on discrete representations or semantic prompts fed into task-specific decoders, which limits the ability of the MLLM to capture fine-grained visual details. To address these challenges, we introduce a segmentation framework for MLLM based on image generation, which naturally produces dense masks for target objects. We leverage MLLM to output visual tokens and detokenize them into images using an universal VQ-VAE, making the segmentation fully dependent on the pixel-level understanding of the MLLM. To reduce inference latency, we employ a next-scale-prediction strategy to generate required visual tokens in parallel. Extensive experiments demonstrate that our method surpasses prior state-of-the-art approaches on multiple segmentation datasets with a remarkable boost in inference speed, while maintaining strong understanding capabilities.
PruneHal: Reducing Hallucinations in Multi-modal Large Language Models through Adaptive KV Cache Pruning
Oct 22, 2025Abstract:While multi-modal large language models (MLLMs) have made significant progress in recent years, the issue of hallucinations remains a major challenge. To mitigate this phenomenon, existing solutions either introduce additional data for further training or incorporate external or internal information during inference. However, these approaches inevitably introduce extra computational costs. In this paper, we observe that hallucinations in MLLMs are strongly associated with insufficient attention allocated to visual tokens. In particular, the presence of redundant visual tokens disperses the model's attention, preventing it from focusing on the most informative ones. As a result, critical visual cues are often under-attended, which in turn exacerbates the occurrence of hallucinations. Building on this observation, we propose \textbf{PruneHal}, a training-free, simple yet effective method that leverages adaptive KV cache pruning to enhance the model's focus on critical visual information, thereby mitigating hallucinations. To the best of our knowledge, we are the first to apply token pruning for hallucination mitigation in MLLMs. Notably, our method don't require additional training and incurs nearly no extra inference cost. Moreover, PruneHal is model-agnostic and can be seamlessly integrated with different decoding strategies, including those specifically designed for hallucination mitigation. We evaluate PruneHal on several widely used hallucination evaluation benchmarks using four mainstream MLLMs, achieving robust and outstanding results that highlight the effectiveness and superiority of our method. Our code will be publicly available.
Ming-Omni: A Unified Multimodal Model for Perception and Generation
Jun 11, 2025


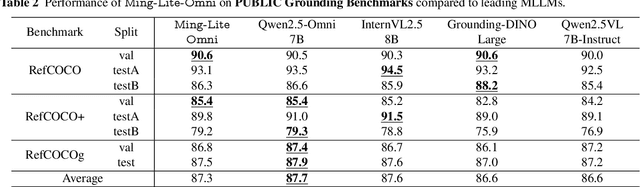
Abstract:We propose Ming-Omni, a unified multimodal model capable of processing images, text, audio, and video, while demonstrating strong proficiency in both speech and image generation. Ming-Omni employs dedicated encoders to extract tokens from different modalities, which are then processed by Ling, an MoE architecture equipped with newly proposed modality-specific routers. This design enables a single model to efficiently process and fuse multimodal inputs within a unified framework, thereby facilitating diverse tasks without requiring separate models, task-specific fine-tuning, or structural redesign. Importantly, Ming-Omni extends beyond conventional multimodal models by supporting audio and image generation. This is achieved through the integration of an advanced audio decoder for natural-sounding speech and Ming-Lite-Uni for high-quality image generation, which also allow the model to engage in context-aware chatting, perform text-to-speech conversion, and conduct versatile image editing. Our experimental results showcase Ming-Omni offers a powerful solution for unified perception and generation across all modalities. Notably, our proposed Ming-Omni is the first open-source model we are aware of to match GPT-4o in modality support, and we release all code and model weights to encourage further research and development in the community.
Ming-Lite-Uni: Advancements in Unified Architecture for Natural Multimodal Interaction
May 05, 2025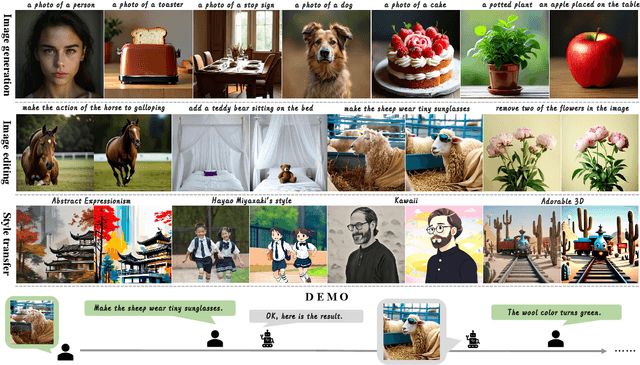
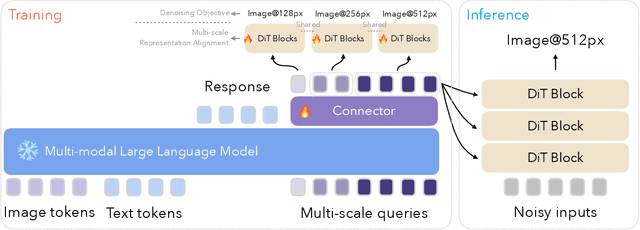
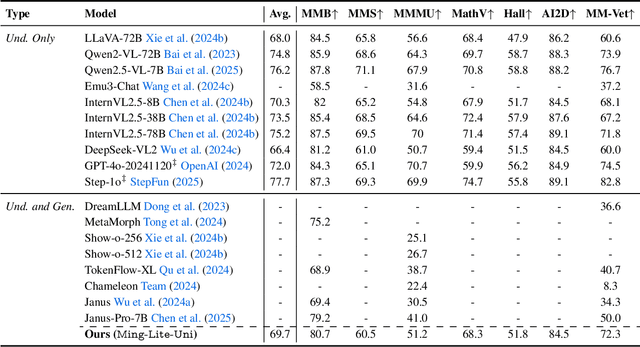
Abstract:We introduce Ming-Lite-Uni, an open-source multimodal framework featuring a newly designed unified visual generator and a native multimodal autoregressive model tailored for unifying vision and language. Specifically, this project provides an open-source implementation of the integrated MetaQueries and M2-omni framework, while introducing the novel multi-scale learnable tokens and multi-scale representation alignment strategy. By leveraging a fixed MLLM and a learnable diffusion model, Ming-Lite-Uni enables native multimodal AR models to perform both text-to-image generation and instruction based image editing tasks, expanding their capabilities beyond pure visual understanding. Our experimental results demonstrate the strong performance of Ming-Lite-Uni and illustrate the impressive fluid nature of its interactive process. All code and model weights are open-sourced to foster further exploration within the community. Notably, this work aligns with concurrent multimodal AI milestones - such as ChatGPT-4o with native image generation updated in March 25, 2025 - underscoring the broader significance of unified models like Ming-Lite-Uni on the path toward AGI. Ming-Lite-Uni is in alpha stage and will soon be further refined.
A Continual Learning-driven Model for Accurate and Generalizable Segmentation of Clinically Comprehensive and Fine-grained Whole-body Anatomies in CT
Mar 16, 2025Abstract:Precision medicine in the quantitative management of chronic diseases and oncology would be greatly improved if the Computed Tomography (CT) scan of any patient could be segmented, parsed and analyzed in a precise and detailed way. However, there is no such fully annotated CT dataset with all anatomies delineated for training because of the exceptionally high manual cost, the need for specialized clinical expertise, and the time required to finish the task. To this end, we proposed a novel continual learning-driven CT model that can segment complete anatomies presented using dozens of previously partially labeled datasets, dynamically expanding its capacity to segment new ones without compromising previously learned organ knowledge. Existing multi-dataset approaches are not able to dynamically segment new anatomies without catastrophic forgetting and would encounter optimization difficulty or infeasibility when segmenting hundreds of anatomies across the whole range of body regions. Our single unified CT segmentation model, CL-Net, can highly accurately segment a clinically comprehensive set of 235 fine-grained whole-body anatomies. Composed of a universal encoder, multiple optimized and pruned decoders, CL-Net is developed using 13,952 CT scans from 20 public and 16 private high-quality partially labeled CT datasets of various vendors, different contrast phases, and pathologies. Extensive evaluation demonstrates that CL-Net consistently outperforms the upper limit of an ensemble of 36 specialist nnUNets trained per dataset with the complexity of 5% model size and significantly surpasses the segmentation accuracy of recent leading Segment Anything-style medical image foundation models by large margins. Our continual learning-driven CL-Net model would lay a solid foundation to facilitate many downstream tasks of oncology and chronic diseases using the most widely adopted CT imaging.
From Slices to Sequences: Autoregressive Tracking Transformer for Cohesive and Consistent 3D Lymph Node Detection in CT Scans
Mar 11, 2025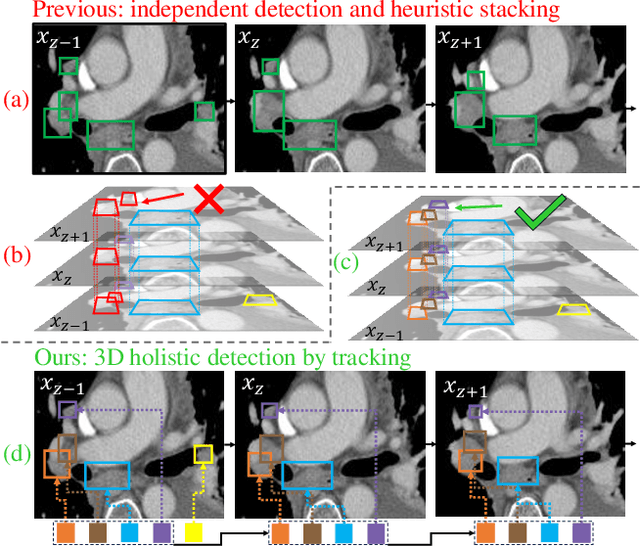

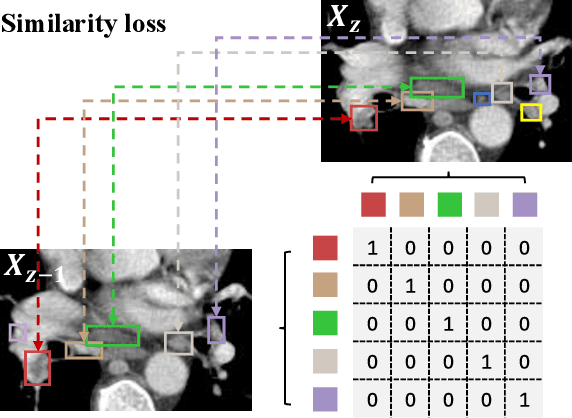
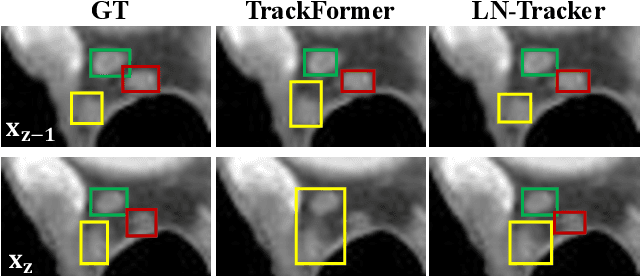
Abstract:Lymph node (LN) assessment is an essential task in the routine radiology workflow, providing valuable insights for cancer staging, treatment planning and beyond. Identifying scatteredly-distributed and low-contrast LNs in 3D CT scans is highly challenging, even for experienced clinicians. Previous lesion and LN detection methods demonstrate effectiveness of 2.5D approaches (i.e, using 2D network with multi-slice inputs), leveraging pretrained 2D model weights and showing improved accuracy as compared to separate 2D or 3D detectors. However, slice-based 2.5D detectors do not explicitly model inter-slice consistency for LN as a 3D object, requiring heuristic post-merging steps to generate final 3D LN instances, which can involve tuning a set of parameters for each dataset. In this work, we formulate 3D LN detection as a tracking task and propose LN-Tracker, a novel LN tracking transformer, for joint end-to-end detection and 3D instance association. Built upon DETR-based detector, LN-Tracker decouples transformer decoder's query into the track and detection groups, where the track query autoregressively follows previously tracked LN instances along the z-axis of a CT scan. We design a new transformer decoder with masked attention module to align track query's content to the context of current slice, meanwhile preserving detection query's high accuracy in current slice. An inter-slice similarity loss is introduced to encourage cohesive LN association between slices. Extensive evaluation on four lymph node datasets shows LN-Tracker's superior performance, with at least 2.7% gain in average sensitivity when compared to other top 3D/2.5D detectors. Further validation on public lung nodule and prostate tumor detection tasks confirms the generalizability of LN-Tracker as it achieves top performance on both tasks. Datasets will be released upon acceptance.
MotionStone: Decoupled Motion Intensity Modulation with Diffusion Transformer for Image-to-Video Generation
Dec 08, 2024



Abstract:The image-to-video (I2V) generation is conditioned on the static image, which has been enhanced recently by the motion intensity as an additional control signal. These motion-aware models are appealing to generate diverse motion patterns, yet there lacks a reliable motion estimator for training such models on large-scale video set in the wild. Traditional metrics, e.g., SSIM or optical flow, are hard to generalize to arbitrary videos, while, it is very tough for human annotators to label the abstract motion intensity neither. Furthermore, the motion intensity shall reveal both local object motion and global camera movement, which has not been studied before. This paper addresses the challenge with a new motion estimator, capable of measuring the decoupled motion intensities of objects and cameras in video. We leverage the contrastive learning on randomly paired videos and distinguish the video with greater motion intensity. Such a paradigm is friendly for annotation and easy to scale up to achieve stable performance on motion estimation. We then present a new I2V model, named MotionStone, developed with the decoupled motion estimator. Experimental results demonstrate the stability of the proposed motion estimator and the state-of-the-art performance of MotionStone on I2V generation. These advantages warrant the decoupled motion estimator to serve as a general plug-in enhancer for both data processing and video generation training.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge