Yirui Wang
EMMA: Generalizing Real-World Robot Manipulation via Generative Visual Transfer
Sep 26, 2025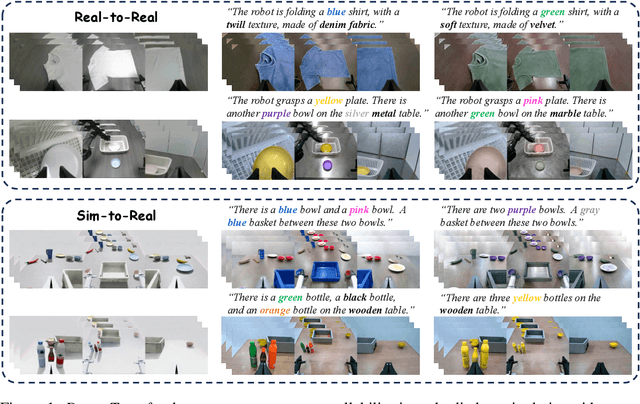
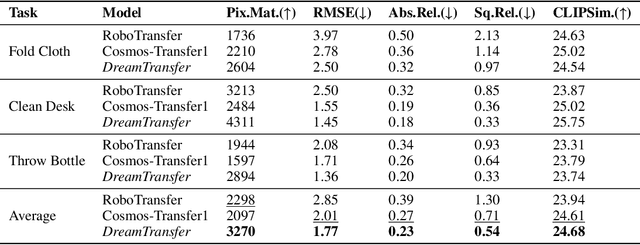
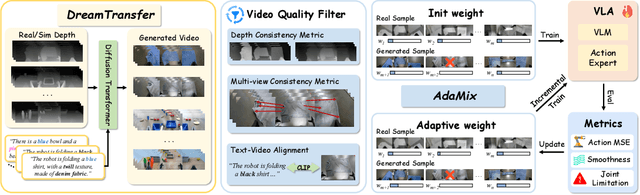
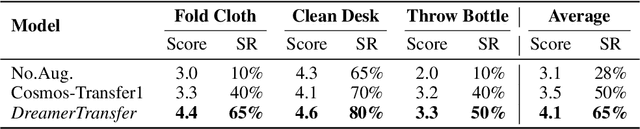
Abstract:Vision-language-action (VLA) models increasingly rely on diverse training data to achieve robust generalization. However, collecting large-scale real-world robot manipulation data across varied object appearances and environmental conditions remains prohibitively time-consuming and expensive. To overcome this bottleneck, we propose Embodied Manipulation Media Adaptation (EMMA), a VLA policy enhancement framework that integrates a generative data engine with an effective training pipeline. We introduce DreamTransfer, a diffusion Transformer-based framework for generating multi-view consistent, geometrically grounded embodied manipulation videos. DreamTransfer enables text-controlled visual editing of robot videos, transforming foreground, background, and lighting conditions without compromising 3D structure or geometrical plausibility. Furthermore, we explore hybrid training with real and generated data, and introduce AdaMix, a hard-sample-aware training strategy that dynamically reweights training batches to focus optimization on perceptually or kinematically challenging samples. Extensive experiments show that videos generated by DreamTransfer significantly outperform prior video generation methods in multi-view consistency, geometric fidelity, and text-conditioning accuracy. Crucially, VLAs trained with generated data enable robots to generalize to unseen object categories and novel visual domains using only demonstrations from a single appearance. In real-world robotic manipulation tasks with zero-shot visual domains, our approach achieves over a 200% relative performance gain compared to training on real data alone, and further improves by 13% with AdaMix, demonstrating its effectiveness in boosting policy generalization.
Pseudo Depth Meets Gaussian: A Feed-forward RGB SLAM Baseline
Aug 06, 2025Abstract:Incrementally recovering real-sized 3D geometry from a pose-free RGB stream is a challenging task in 3D reconstruction, requiring minimal assumptions on input data. Existing methods can be broadly categorized into end-to-end and visual SLAM-based approaches, both of which either struggle with long sequences or depend on slow test-time optimization and depth sensors. To address this, we first integrate a depth estimator into an RGB-D SLAM system, but this approach is hindered by inaccurate geometric details in predicted depth. Through further investigation, we find that 3D Gaussian mapping can effectively solve this problem. Building on this, we propose an online 3D reconstruction method using 3D Gaussian-based SLAM, combined with a feed-forward recurrent prediction module to directly infer camera pose from optical flow. This approach replaces slow test-time optimization with fast network inference, significantly improving tracking speed. Additionally, we introduce a local graph rendering technique to enhance robustness in feed-forward pose prediction. Experimental results on the Replica and TUM-RGBD datasets, along with a real-world deployment demonstration, show that our method achieves performance on par with the state-of-the-art SplaTAM, while reducing tracking time by more than 90\%.
A Continual Learning-driven Model for Accurate and Generalizable Segmentation of Clinically Comprehensive and Fine-grained Whole-body Anatomies in CT
Mar 16, 2025Abstract:Precision medicine in the quantitative management of chronic diseases and oncology would be greatly improved if the Computed Tomography (CT) scan of any patient could be segmented, parsed and analyzed in a precise and detailed way. However, there is no such fully annotated CT dataset with all anatomies delineated for training because of the exceptionally high manual cost, the need for specialized clinical expertise, and the time required to finish the task. To this end, we proposed a novel continual learning-driven CT model that can segment complete anatomies presented using dozens of previously partially labeled datasets, dynamically expanding its capacity to segment new ones without compromising previously learned organ knowledge. Existing multi-dataset approaches are not able to dynamically segment new anatomies without catastrophic forgetting and would encounter optimization difficulty or infeasibility when segmenting hundreds of anatomies across the whole range of body regions. Our single unified CT segmentation model, CL-Net, can highly accurately segment a clinically comprehensive set of 235 fine-grained whole-body anatomies. Composed of a universal encoder, multiple optimized and pruned decoders, CL-Net is developed using 13,952 CT scans from 20 public and 16 private high-quality partially labeled CT datasets of various vendors, different contrast phases, and pathologies. Extensive evaluation demonstrates that CL-Net consistently outperforms the upper limit of an ensemble of 36 specialist nnUNets trained per dataset with the complexity of 5% model size and significantly surpasses the segmentation accuracy of recent leading Segment Anything-style medical image foundation models by large margins. Our continual learning-driven CL-Net model would lay a solid foundation to facilitate many downstream tasks of oncology and chronic diseases using the most widely adopted CT imaging.
From Slices to Sequences: Autoregressive Tracking Transformer for Cohesive and Consistent 3D Lymph Node Detection in CT Scans
Mar 11, 2025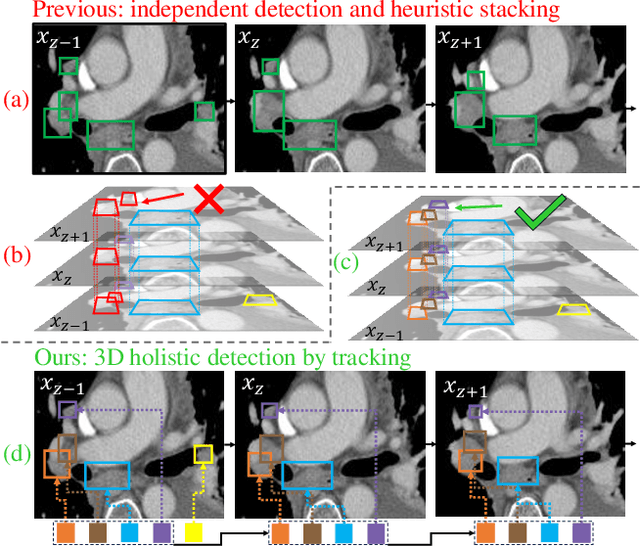

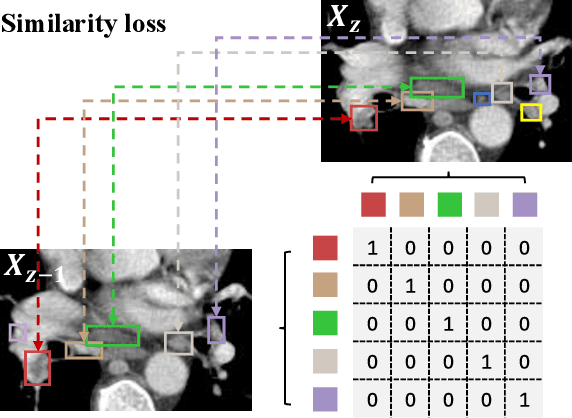
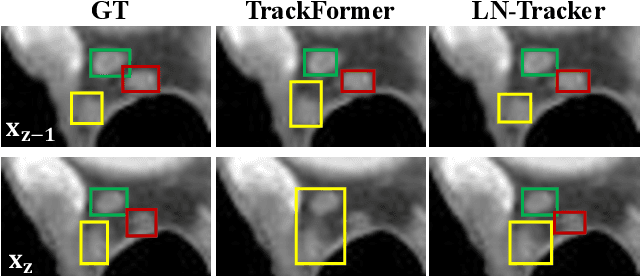
Abstract:Lymph node (LN) assessment is an essential task in the routine radiology workflow, providing valuable insights for cancer staging, treatment planning and beyond. Identifying scatteredly-distributed and low-contrast LNs in 3D CT scans is highly challenging, even for experienced clinicians. Previous lesion and LN detection methods demonstrate effectiveness of 2.5D approaches (i.e, using 2D network with multi-slice inputs), leveraging pretrained 2D model weights and showing improved accuracy as compared to separate 2D or 3D detectors. However, slice-based 2.5D detectors do not explicitly model inter-slice consistency for LN as a 3D object, requiring heuristic post-merging steps to generate final 3D LN instances, which can involve tuning a set of parameters for each dataset. In this work, we formulate 3D LN detection as a tracking task and propose LN-Tracker, a novel LN tracking transformer, for joint end-to-end detection and 3D instance association. Built upon DETR-based detector, LN-Tracker decouples transformer decoder's query into the track and detection groups, where the track query autoregressively follows previously tracked LN instances along the z-axis of a CT scan. We design a new transformer decoder with masked attention module to align track query's content to the context of current slice, meanwhile preserving detection query's high accuracy in current slice. An inter-slice similarity loss is introduced to encourage cohesive LN association between slices. Extensive evaluation on four lymph node datasets shows LN-Tracker's superior performance, with at least 2.7% gain in average sensitivity when compared to other top 3D/2.5D detectors. Further validation on public lung nodule and prostate tumor detection tasks confirms the generalizability of LN-Tracker as it achieves top performance on both tasks. Datasets will be released upon acceptance.
Leveraging Semantic Asymmetry for Precise Gross Tumor Volume Segmentation of Nasopharyngeal Carcinoma in Planning CT
Nov 27, 2024
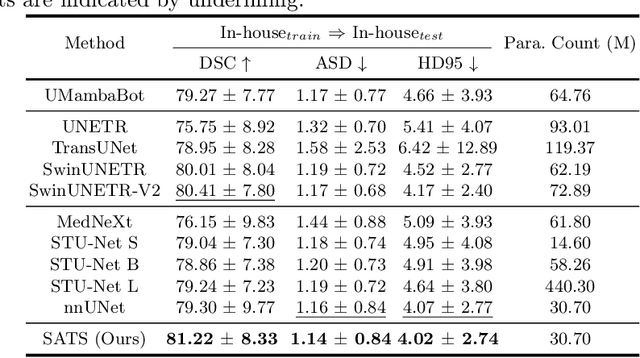
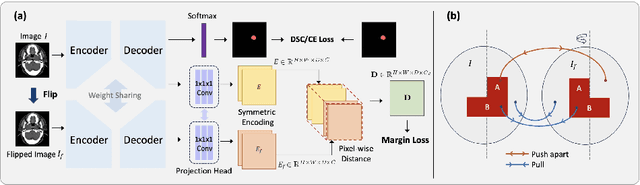
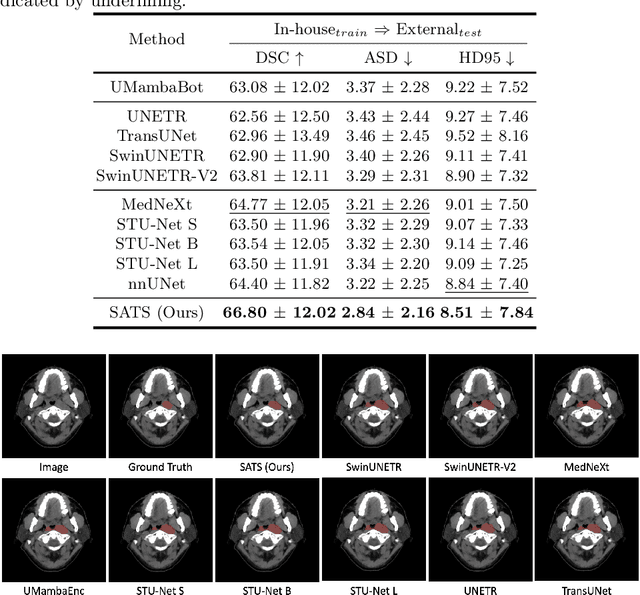
Abstract:In the radiation therapy of nasopharyngeal carcinoma (NPC), clinicians typically delineate the gross tumor volume (GTV) using non-contrast planning computed tomography to ensure accurate radiation dose delivery. However, the low contrast between tumors and adjacent normal tissues necessitates that radiation oncologists manually delineate the tumors, often relying on diagnostic MRI for guidance. % In this study, we propose a novel approach to directly segment NPC gross tumors on non-contrast planning CT images, circumventing potential registration errors when aligning MRI or MRI-derived tumor masks to planning CT. To address the low contrast issues between tumors and adjacent normal structures in planning CT, we introduce a 3D Semantic Asymmetry Tumor segmentation (SATs) method. Specifically, we posit that a healthy nasopharyngeal region is characteristically bilaterally symmetric, whereas the emergence of nasopharyngeal carcinoma disrupts this symmetry. Then, we propose a Siamese contrastive learning segmentation framework that minimizes the voxel-wise distance between original and flipped areas without tumor and encourages a larger distance between original and flipped areas with tumor. Thus, our approach enhances the sensitivity of features to semantic asymmetries. % Extensive experiments demonstrate that the proposed SATs achieves the leading NPC GTV segmentation performance in both internal and external testing, \emph{e.g.}, with at least 2\% absolute Dice score improvement and 12\% average distance error reduction when compared to other state-of-the-art methods in the external testing.
Effective Lymph Nodes Detection in CT Scans Using Location Debiased Query Selection and Contrastive Query Representation in Transformer
Apr 04, 2024Abstract:Lymph node (LN) assessment is a critical, indispensable yet very challenging task in the routine clinical workflow of radiology and oncology. Accurate LN analysis is essential for cancer diagnosis, staging, and treatment planning. Finding scatteredly distributed, low-contrast clinically relevant LNs in 3D CT is difficult even for experienced physicians under high inter-observer variations. Previous automatic LN detection works typically yield limited recall and high false positives (FPs) due to adjacent anatomies with similar image intensities, shapes, or textures (vessels, muscles, esophagus, etc). In this work, we propose a new LN DEtection TRansformer, named LN-DETR, to achieve more accurate performance. By enhancing the 2D backbone with a multi-scale 2.5D feature fusion to incorporate 3D context explicitly, more importantly, we make two main contributions to improve the representation quality of LN queries. 1) Considering that LN boundaries are often unclear, an IoU prediction head and a location debiased query selection are proposed to select LN queries of higher localization accuracy as the decoder query's initialization. 2) To reduce FPs, query contrastive learning is employed to explicitly reinforce LN queries towards their best-matched ground-truth queries over unmatched query predictions. Trained and tested on 3D CT scans of 1067 patients (with 10,000+ labeled LNs) via combining seven LN datasets from different body parts (neck, chest, and abdomen) and pathologies/cancers, our method significantly improves the performance of previous leading methods by > 4-5% average recall at the same FP rates in both internal and external testing. We further evaluate on the universal lesion detection task using NIH DeepLesion benchmark, and our method achieves the top performance of 88.46% averaged recall across 0.5 to 4 FPs per image, compared with other leading reported results.
Improving and Benchmarking Offline Reinforcement Learning Algorithms
Jun 01, 2023Abstract:Recently, Offline Reinforcement Learning (RL) has achieved remarkable progress with the emergence of various algorithms and datasets. However, these methods usually focus on algorithmic advancements, ignoring that many low-level implementation choices considerably influence or even drive the final performance. As a result, it becomes hard to attribute the progress in Offline RL as these choices are not sufficiently discussed and aligned in the literature. In addition, papers focusing on a dataset (e.g., D4RL) often ignore algorithms proposed on another dataset (e.g., RL Unplugged), causing isolation among the algorithms, which might slow down the overall progress. Therefore, this work aims to bridge the gaps caused by low-level choices and datasets. To this end, we empirically investigate 20 implementation choices using three representative algorithms (i.e., CQL, CRR, and IQL) and present a guidebook for choosing implementations. Following the guidebook, we find two variants CRR+ and CQL+ , achieving new state-of-the-art on D4RL. Moreover, we benchmark eight popular offline RL algorithms across datasets under unified training and evaluation framework. The findings are inspiring: the success of a learning paradigm severely depends on the data distribution, and some previous conclusions are biased by the dataset used. Our code is available at https://github.com/sail-sg/offbench.
Accurate and Robust Lesion RECIST Diameter Prediction and Segmentation with Transformers
Aug 28, 2022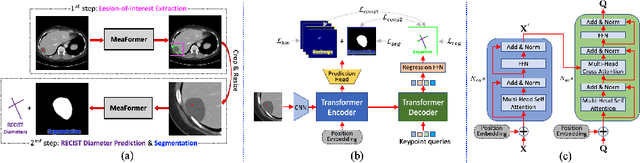
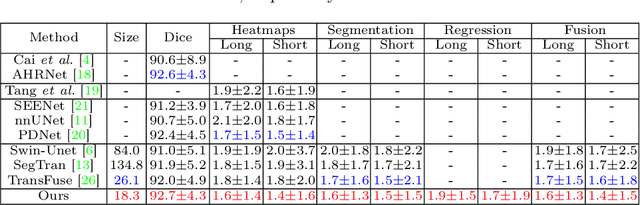
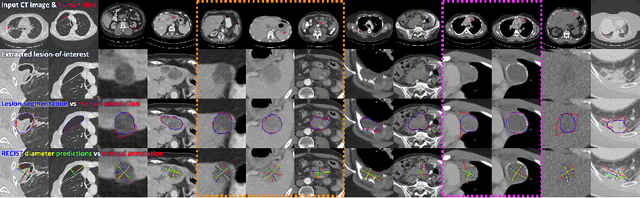
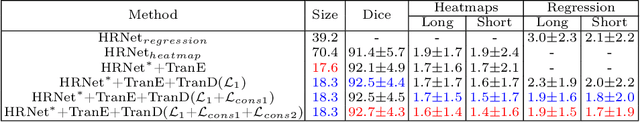
Abstract:Automatically measuring lesion/tumor size with RECIST (Response Evaluation Criteria In Solid Tumors) diameters and segmentation is important for computer-aided diagnosis. Although it has been studied in recent years, there is still space to improve its accuracy and robustness, such as (1) enhancing features by incorporating rich contextual information while keeping a high spatial resolution and (2) involving new tasks and losses for joint optimization. To reach this goal, this paper proposes a transformer-based network (MeaFormer, Measurement transFormer) for lesion RECIST diameter prediction and segmentation (LRDPS). It is formulated as three correlative and complementary tasks: lesion segmentation, heatmap prediction, and keypoint regression. To the best of our knowledge, it is the first time to use keypoint regression for RECIST diameter prediction. MeaFormer can enhance high-resolution features by employing transformers to capture their long-range dependencies. Two consistency losses are introduced to explicitly build relationships among these tasks for better optimization. Experiments show that MeaFormer achieves the state-of-the-art performance of LRDPS on the large-scale DeepLesion dataset and produces promising results of two downstream clinic-relevant tasks, i.e., 3D lesion segmentation and RECIST assessment in longitudinal studies.
PieTrack: An MOT solution based on synthetic data training and self-supervised domain adaptation
Jul 22, 2022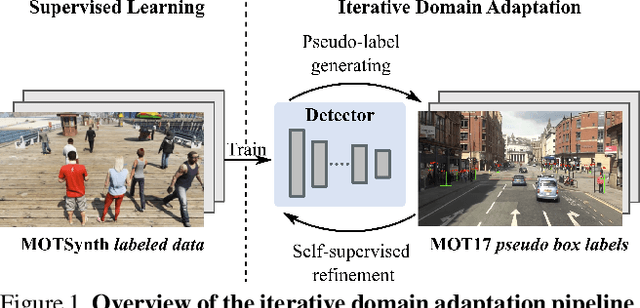
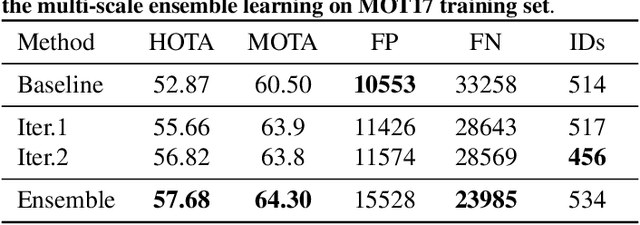
Abstract:In order to cope with the increasing demand for labeling data and privacy issues with human detection, synthetic data has been used as a substitute and showing promising results in human detection and tracking tasks. We participate in the 7th Workshop on Benchmarking Multi-Target Tracking (BMTT), themed on "How Far Can Synthetic Data Take us"? Our solution, PieTrack, is developed based on synthetic data without using any pre-trained weights. We propose a self-supervised domain adaptation method that enables mitigating the domain shift issue between the synthetic (e.g., MOTSynth) and real data (e.g., MOT17) without involving extra human labels. By leveraging the proposed multi-scale ensemble inference, we achieved a final HOTA score of 58.7 on the MOT17 testing set, ranked third place in the challenge.
Coherence Learning using Keypoint-based Pooling Network for Accurately Assessing Radiographic Knee Osteoarthritis
Dec 16, 2021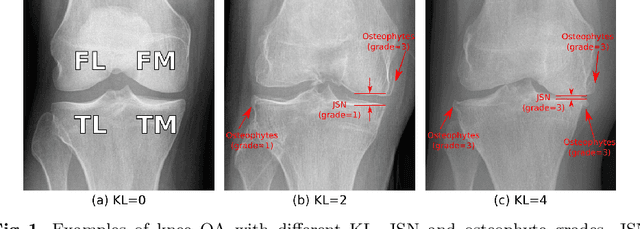
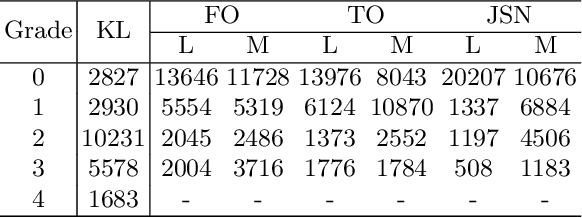
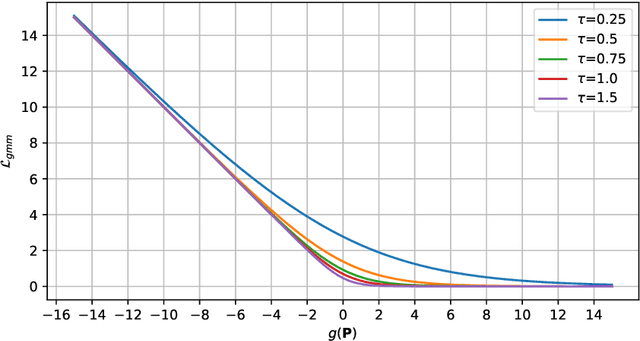
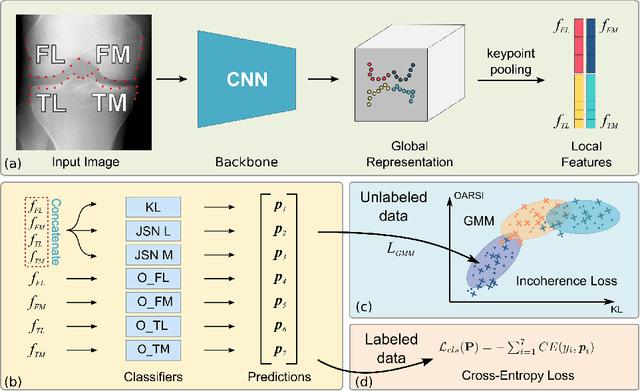
Abstract:Knee osteoarthritis (OA) is a common degenerate joint disorder that affects a large population of elderly people worldwide. Accurate radiographic assessment of knee OA severity plays a critical role in chronic patient management. Current clinically-adopted knee OA grading systems are observer subjective and suffer from inter-rater disagreements. In this work, we propose a computer-aided diagnosis approach to provide more accurate and consistent assessments of both composite and fine-grained OA grades simultaneously. A novel semi-supervised learning method is presented to exploit the underlying coherence in the composite and fine-grained OA grades by learning from unlabeled data. By representing the grade coherence using the log-probability of a pre-trained Gaussian Mixture Model, we formulate an incoherence loss to incorporate unlabeled data in training. The proposed method also describes a keypoint-based pooling network, where deep image features are pooled from the disease-targeted keypoints (extracted along the knee joint) to provide more aligned and pathologically informative feature representations, for accurate OA grade assessments. The proposed method is comprehensively evaluated on the public Osteoarthritis Initiative (OAI) data, a multi-center ten-year observational study on 4,796 subjects. Experimental results demonstrate that our method leads to significant improvements over previous strong whole image-based deep classification network baselines (like ResNet-50).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge