Chang-Fu Kuo
oRetrieval Augmented Generation for 10 Large Language Models and its Generalizability in Assessing Medical Fitness
Oct 11, 2024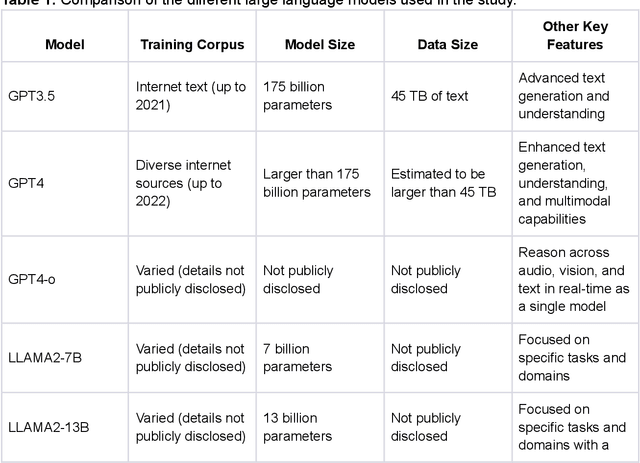



Abstract:Large Language Models (LLMs) show potential for medical applications but often lack specialized clinical knowledge. Retrieval Augmented Generation (RAG) allows customization with domain-specific information, making it suitable for healthcare. This study evaluates the accuracy, consistency, and safety of RAG models in determining fitness for surgery and providing preoperative instructions. We developed LLM-RAG models using 35 local and 23 international preoperative guidelines and tested them against human-generated responses. A total of 3,682 responses were evaluated. Clinical documents were processed using Llamaindex, and 10 LLMs, including GPT3.5, GPT4, and Claude-3, were assessed. Fourteen clinical scenarios were analyzed, focusing on seven aspects of preoperative instructions. Established guidelines and expert judgment were used to determine correct responses, with human-generated answers serving as comparisons. The LLM-RAG models generated responses within 20 seconds, significantly faster than clinicians (10 minutes). The GPT4 LLM-RAG model achieved the highest accuracy (96.4% vs. 86.6%, p=0.016), with no hallucinations and producing correct instructions comparable to clinicians. Results were consistent across both local and international guidelines. This study demonstrates the potential of LLM-RAG models for preoperative healthcare tasks, highlighting their efficiency, scalability, and reliability.
Lumbar Bone Mineral Density Estimation from Chest X-ray Images: Anatomy-aware Attentive Multi-ROI Modeling
Jan 05, 2022



Abstract:Osteoporosis is a common chronic metabolic bone disease that is often under-diagnosed and under-treated due to the limited access to bone mineral density (BMD) examinations, e.g. via Dual-energy X-ray Absorptiometry (DXA). In this paper, we propose a method to predict BMD from Chest X-ray (CXR), one of the most commonly accessible and low-cost medical imaging examinations. Our method first automatically detects Regions of Interest (ROIs) of local and global bone structures from the CXR. Then a multi-ROI deep model with transformer encoder is developed to exploit both local and global information in the chest X-ray image for accurate BMD estimation. Our method is evaluated on 13719 CXR patient cases with their ground truth BMD scores measured by gold-standard DXA. The model predicted BMD has a strong correlation with the ground truth (Pearson correlation coefficient 0.889 on lumbar 1). When applied for osteoporosis screening, it achieves a high classification performance (AUC 0.963 on lumbar 1). As the first effort in the field using CXR scans to predict the BMD, the proposed algorithm holds strong potential in early osteoporosis screening and public health promotion.
Coherence Learning using Keypoint-based Pooling Network for Accurately Assessing Radiographic Knee Osteoarthritis
Dec 16, 2021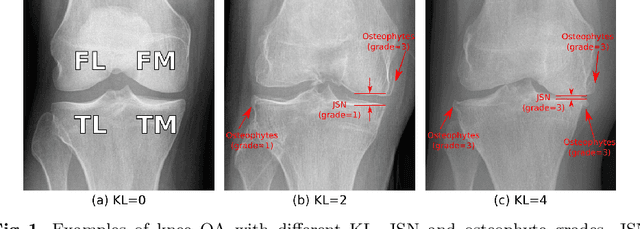
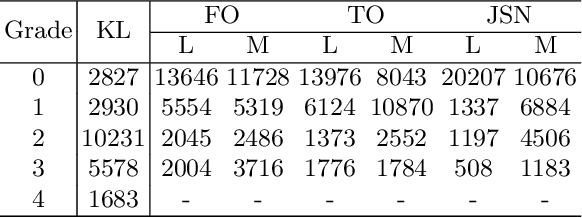
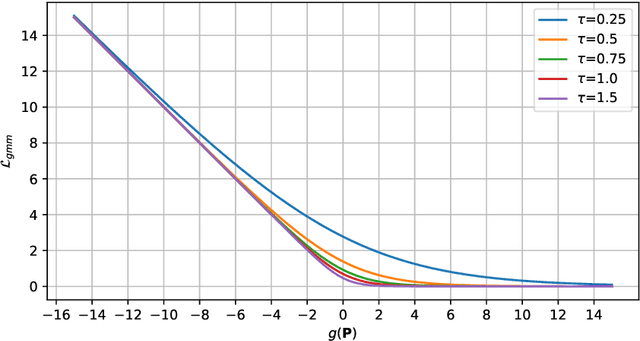
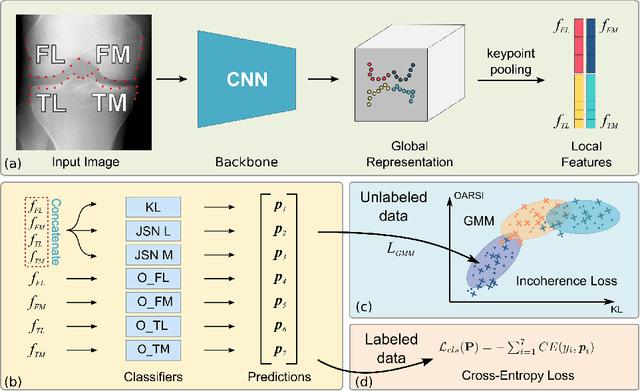
Abstract:Knee osteoarthritis (OA) is a common degenerate joint disorder that affects a large population of elderly people worldwide. Accurate radiographic assessment of knee OA severity plays a critical role in chronic patient management. Current clinically-adopted knee OA grading systems are observer subjective and suffer from inter-rater disagreements. In this work, we propose a computer-aided diagnosis approach to provide more accurate and consistent assessments of both composite and fine-grained OA grades simultaneously. A novel semi-supervised learning method is presented to exploit the underlying coherence in the composite and fine-grained OA grades by learning from unlabeled data. By representing the grade coherence using the log-probability of a pre-trained Gaussian Mixture Model, we formulate an incoherence loss to incorporate unlabeled data in training. The proposed method also describes a keypoint-based pooling network, where deep image features are pooled from the disease-targeted keypoints (extracted along the knee joint) to provide more aligned and pathologically informative feature representations, for accurate OA grade assessments. The proposed method is comprehensively evaluated on the public Osteoarthritis Initiative (OAI) data, a multi-center ten-year observational study on 4,796 subjects. Experimental results demonstrate that our method leads to significant improvements over previous strong whole image-based deep classification network baselines (like ResNet-50).
Opportunistic Screening of Osteoporosis Using Plain Film Chest X-ray
Apr 05, 2021



Abstract:Osteoporosis is a common chronic metabolic bone disease that is often under-diagnosed and under-treated due to the limited access to bone mineral density (BMD) examinations, Dual-energy X-ray Absorptiometry (DXA). In this paper, we propose a method to predict BMD from Chest X-ray (CXR), one of the most common, accessible, and low-cost medical image examinations. Our method first automatically detects Regions of Interest (ROIs) of local and global bone structures from the CXR. Then a multi-ROI model is developed to exploit both local and global information in the chest X-ray image for accurate BMD estimation. Our method is evaluated on 329 CXR cases with ground truth BMD measured by DXA. The model predicted BMD has a strong correlation with the gold standard DXA BMD (Pearson correlation coefficient 0.840). When applied for osteoporosis screening, it achieves a high classification performance (AUC 0.936). As the first effort in the field to use CXR scans to predict the spine BMD, the proposed algorithm holds strong potential in enabling early osteoporosis screening through routine chest X-rays and contributing to the enhancement of public health.
Semi-Supervised Learning for Bone Mineral Density Estimation in Hip X-ray Images
Mar 24, 2021



Abstract:Bone mineral density (BMD) is a clinically critical indicator of osteoporosis, usually measured by dual-energy X-ray absorptiometry (DEXA). Due to the limited accessibility of DEXA machines and examinations, osteoporosis is often under-diagnosed and under-treated, leading to increased fragility fracture risks. Thus it is highly desirable to obtain BMDs with alternative cost-effective and more accessible medical imaging examinations such as X-ray plain films. In this work, we formulate the BMD estimation from plain hip X-ray images as a regression problem. Specifically, we propose a new semi-supervised self-training algorithm to train the BMD regression model using images coupled with DEXA measured BMDs and unlabeled images with pseudo BMDs. Pseudo BMDs are generated and refined iteratively for unlabeled images during self-training. We also present a novel adaptive triplet loss to improve the model's regression accuracy. On an in-house dataset of 1,090 images (819 unique patients), our BMD estimation method achieves a high Pearson correlation coefficient of 0.8805 to ground-truth BMDs. It offers good feasibility to use the more accessible and cheaper X-ray imaging for opportunistic osteoporosis screening.
Contour Transformer Network for One-shot Segmentation of Anatomical Structures
Dec 02, 2020



Abstract:Accurate segmentation of anatomical structures is vital for medical image analysis. The state-of-the-art accuracy is typically achieved by supervised learning methods, where gathering the requisite expert-labeled image annotations in a scalable manner remains a main obstacle. Therefore, annotation-efficient methods that permit to produce accurate anatomical structure segmentation are highly desirable. In this work, we present Contour Transformer Network (CTN), a one-shot anatomy segmentation method with a naturally built-in human-in-the-loop mechanism. We formulate anatomy segmentation as a contour evolution process and model the evolution behavior by graph convolutional networks (GCNs). Training the CTN model requires only one labeled image exemplar and leverages additional unlabeled data through newly introduced loss functions that measure the global shape and appearance consistency of contours. On segmentation tasks of four different anatomies, we demonstrate that our one-shot learning method significantly outperforms non-learning-based methods and performs competitively to the state-of-the-art fully supervised deep learning methods. With minimal human-in-the-loop editing feedback, the segmentation performance can be further improved to surpass the fully supervised methods.
Learning to Segment Anatomical Structures Accurately from One Exemplar
Jul 08, 2020



Abstract:Accurate segmentation of critical anatomical structures is at the core of medical image analysis. The main bottleneck lies in gathering the requisite expert-labeled image annotations in a scalable manner. Methods that permit to produce accurate anatomical structure segmentation without using a large amount of fully annotated training images are highly desirable. In this work, we propose a novel contribution of Contour Transformer Network (CTN), a one-shot anatomy segmentor including a naturally built-in human-in-the-loop mechanism. Segmentation is formulated by learning a contour evolution behavior process based on graph convolutional networks (GCNs). Training of our CTN model requires only one labeled image exemplar and leverages additional unlabeled data through newly introduced loss functions that measure the global shape and appearance consistency of contours. We demonstrate that our one-shot learning method significantly outperforms non-learning-based methods and performs competitively to the state-of-the-art fully supervised deep learning approaches. With minimal human-in-the-loop editing feedback, the segmentation performance can be further improved and tailored towards the observer desired outcomes. This can facilitate the clinician designed imaging-based biomarker assessments (to support personalized quantitative clinical diagnosis) and outperforms fully supervised baselines.
Structured Landmark Detection via Topology-Adapting Deep Graph Learning
Apr 23, 2020



Abstract:Image landmark detection aims to automatically identify the locations of predefined fiducial points. Despite recent success in this filed, higher-ordered structural modeling to capture implicit or explicit relationships among anatomical landmarks has not been adequately exploited. In this work, we present a new topology-adapting deep graph learning approach for accurate anatomical facial and medical (e.g., hand, pelvis) landmark detection. The proposed method constructs graph signals leveraging both local image features and global shape features. The adaptive graph topology naturally explores and lands on task-specific structures which is learned end-to-end with two Graph Convolutional Networks (GCNs). Extensive experiments are conducted on three public facial image datasets (WFLW, 300W and COFW-68) as well as three real-world X-ray medical datasets (Cephalometric (public), Hand and Pelvis). Quantitative results comparing with the previous state-of-the-art approaches across all studied datasets indicating the superior performance in both robustness and accuracy. Qualitative visualizations of the learned graph topologies demonstrate a physically plausible connectivity laying behind the landmarks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge