Kabilan Elangovan
Clinical Validation of Medical-based Large Language Model Chatbots on Ophthalmic Patient Queries with LLM-based Evaluation
Feb 05, 2026Abstract:Domain specific large language models are increasingly used to support patient education, triage, and clinical decision making in ophthalmology, making rigorous evaluation essential to ensure safety and accuracy. This study evaluated four small medical LLMs Meerkat-7B, BioMistral-7B, OpenBioLLM-8B, and MedLLaMA3-v20 in answering ophthalmology related patient queries and assessed the feasibility of LLM based evaluation against clinician grading. In this cross sectional study, 180 ophthalmology patient queries were answered by each model, generating 2160 responses. Models were selected for parameter sizes under 10 billion to enable resource efficient deployment. Responses were evaluated by three ophthalmologists of differing seniority and by GPT-4-Turbo using the S.C.O.R.E. framework assessing safety, consensus and context, objectivity, reproducibility, and explainability, with ratings assigned on a five point Likert scale. Agreement between LLM and clinician grading was assessed using Spearman rank correlation, Kendall tau statistics, and kernel density estimate analyses. Meerkat-7B achieved the highest performance with mean scores of 3.44 from Senior Consultants, 4.08 from Consultants, and 4.18 from Residents. MedLLaMA3-v20 performed poorest, with 25.5 percent of responses containing hallucinations or clinically misleading content, including fabricated terminology. GPT-4-Turbo grading showed strong alignment with clinician assessments overall, with Spearman rho of 0.80 and Kendall tau of 0.67, though Senior Consultants graded more conservatively. Overall, medical LLMs demonstrated potential for safe ophthalmic question answering, but gaps remained in clinical depth and consensus, supporting the feasibility of LLM based evaluation for large scale benchmarking and the need for hybrid automated and clinician review frameworks to guide safe clinical deployment.
Real-world Deployment and Evaluation of PErioperative AI CHatbot (PEACH) -- a Large Language Model Chatbot for Perioperative Medicine
Dec 24, 2024



Abstract:Large Language Models (LLMs) are emerging as powerful tools in healthcare, particularly for complex, domain-specific tasks. This study describes the development and evaluation of the PErioperative AI CHatbot (PEACH), a secure LLM-based system integrated with local perioperative guidelines to support preoperative clinical decision-making. PEACH was embedded with 35 institutional perioperative protocols in the secure Claude 3.5 Sonet LLM framework within Pair Chat (developed by Singapore Government) and tested in a silent deployment with real-world data. Accuracy, safety, and usability were assessed. Deviations and hallucinations were categorized based on potential harm, and user feedback was evaluated using the Technology Acceptance Model (TAM). Updates were made after the initial silent deployment to amend one protocol. In 240 real-world clinical iterations, PEACH achieved a first-generation accuracy of 97.5% (78/80) and an overall accuracy of 96.7% (232/240) across three iterations. The updated PEACH demonstrated improved accuracy of 97.9% (235/240), with a statistically significant difference from the null hypothesis of 95% accuracy (p = 0.018, 95% CI: 0.952-0.991). Minimal hallucinations and deviations were observed (both 1/240 and 2/240, respectively). Clinicians reported that PEACH expedited decisions in 95% of cases, and inter-rater reliability ranged from kappa 0.772-0.893 within PEACH and 0.610-0.784 among attendings. PEACH is an accurate, adaptable tool that enhances consistency and efficiency in perioperative decision-making. Future research should explore its scalability across specialties and its impact on clinical outcomes.
oRetrieval Augmented Generation for 10 Large Language Models and its Generalizability in Assessing Medical Fitness
Oct 11, 2024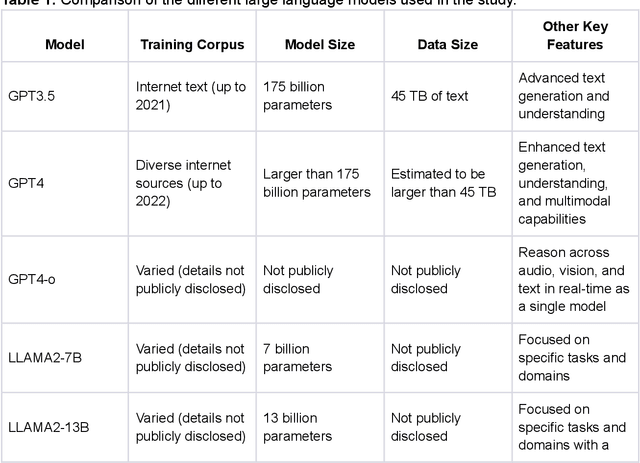



Abstract:Large Language Models (LLMs) show potential for medical applications but often lack specialized clinical knowledge. Retrieval Augmented Generation (RAG) allows customization with domain-specific information, making it suitable for healthcare. This study evaluates the accuracy, consistency, and safety of RAG models in determining fitness for surgery and providing preoperative instructions. We developed LLM-RAG models using 35 local and 23 international preoperative guidelines and tested them against human-generated responses. A total of 3,682 responses were evaluated. Clinical documents were processed using Llamaindex, and 10 LLMs, including GPT3.5, GPT4, and Claude-3, were assessed. Fourteen clinical scenarios were analyzed, focusing on seven aspects of preoperative instructions. Established guidelines and expert judgment were used to determine correct responses, with human-generated answers serving as comparisons. The LLM-RAG models generated responses within 20 seconds, significantly faster than clinicians (10 minutes). The GPT4 LLM-RAG model achieved the highest accuracy (96.4% vs. 86.6%, p=0.016), with no hallucinations and producing correct instructions comparable to clinicians. Results were consistent across both local and international guidelines. This study demonstrates the potential of LLM-RAG models for preoperative healthcare tasks, highlighting their efficiency, scalability, and reliability.
A Proposed S.C.O.R.E. Evaluation Framework for Large Language Models : Safety, Consensus, Objectivity, Reproducibility and Explainability
Jul 10, 2024



Abstract:A comprehensive qualitative evaluation framework for large language models (LLM) in healthcare that expands beyond traditional accuracy and quantitative metrics needed. We propose 5 key aspects for evaluation of LLMs: Safety, Consensus, Objectivity, Reproducibility and Explainability (S.C.O.R.E.). We suggest that S.C.O.R.E. may form the basis for an evaluation framework for future LLM-based models that are safe, reliable, trustworthy, and ethical for healthcare and clinical applications.
Fine-tuning Large Language Model (LLM) Artificial Intelligence Chatbots in Ophthalmology and LLM-based evaluation using GPT-4
Feb 15, 2024



Abstract:Purpose: To assess the alignment of GPT-4-based evaluation to human clinician experts, for the evaluation of responses to ophthalmology-related patient queries generated by fine-tuned LLM chatbots. Methods: 400 ophthalmology questions and paired answers were created by ophthalmologists to represent commonly asked patient questions, divided into fine-tuning (368; 92%), and testing (40; 8%). We find-tuned 5 different LLMs, including LLAMA2-7b, LLAMA2-7b-Chat, LLAMA2-13b, and LLAMA2-13b-Chat. For the testing dataset, additional 8 glaucoma QnA pairs were included. 200 responses to the testing dataset were generated by 5 fine-tuned LLMs for evaluation. A customized clinical evaluation rubric was used to guide GPT-4 evaluation, grounded on clinical accuracy, relevance, patient safety, and ease of understanding. GPT-4 evaluation was then compared against ranking by 5 clinicians for clinical alignment. Results: Among all fine-tuned LLMs, GPT-3.5 scored the highest (87.1%), followed by LLAMA2-13b (80.9%), LLAMA2-13b-chat (75.5%), LLAMA2-7b-Chat (70%) and LLAMA2-7b (68.8%) based on the GPT-4 evaluation. GPT-4 evaluation demonstrated significant agreement with human clinician rankings, with Spearman and Kendall Tau correlation coefficients of 0.90 and 0.80 respectively; while correlation based on Cohen Kappa was more modest at 0.50. Notably, qualitative analysis and the glaucoma sub-analysis revealed clinical inaccuracies in the LLM-generated responses, which were appropriately identified by the GPT-4 evaluation. Conclusion: The notable clinical alignment of GPT-4 evaluation highlighted its potential to streamline the clinical evaluation of LLM chatbot responses to healthcare-related queries. By complementing the existing clinician-dependent manual grading, this efficient and automated evaluation could assist the validation of future developments in LLM applications for healthcare.
Development and Testing of Retrieval Augmented Generation in Large Language Models -- A Case Study Report
Jan 29, 2024Abstract:Purpose: Large Language Models (LLMs) hold significant promise for medical applications. Retrieval Augmented Generation (RAG) emerges as a promising approach for customizing domain knowledge in LLMs. This case study presents the development and evaluation of an LLM-RAG pipeline tailored for healthcare, focusing specifically on preoperative medicine. Methods: We developed an LLM-RAG model using 35 preoperative guidelines and tested it against human-generated responses, with a total of 1260 responses evaluated. The RAG process involved converting clinical documents into text using Python-based frameworks like LangChain and Llamaindex, and processing these texts into chunks for embedding and retrieval. Vector storage techniques and selected embedding models to optimize data retrieval, using Pinecone for vector storage with a dimensionality of 1536 and cosine similarity for loss metrics. Human-generated answers, provided by junior doctors, were used as a comparison. Results: The LLM-RAG model generated answers within an average of 15-20 seconds, significantly faster than the 10 minutes typically required by humans. Among the basic LLMs, GPT4.0 exhibited the best accuracy of 80.1%. This accuracy was further increased to 91.4% when the model was enhanced with RAG. Compared to the human-generated instructions, which had an accuracy of 86.3%, the performance of the GPT4.0 RAG model demonstrated non-inferiority (p=0.610). Conclusions: In this case study, we demonstrated a LLM-RAG model for healthcare implementation. The pipeline shows the advantages of grounded knowledge, upgradability, and scalability as important aspects of healthcare LLM deployment.
Development and Testing of a Novel Large Language Model-Based Clinical Decision Support Systems for Medication Safety in 12 Clinical Specialties
Jan 29, 2024



Abstract:Importance: We introduce a novel Retrieval Augmented Generation (RAG)-Large Language Model (LLM) as a Clinical Decision Support System (CDSS) for safe medication prescription. This model addresses the limitations of traditional rule-based CDSS by providing relevant prescribing error alerts tailored to patient context and institutional guidelines. Objective: The study evaluates the efficacy of an LLM-based CDSS in identifying medication errors across various medical and surgical case vignettes, compared to a human expert panel. It also examines clinician preferences among different CDSS integration modalities: junior pharmacist, LLM-based CDSS alone, and a combination of both. Design, Setting, and Participants: Utilizing a RAG model with GPT-4.0, the study involved 61 prescribing error scenarios within 23 clinical vignettes across 12 specialties. An expert panel assessed these cases using the PCNE classification and NCC MERP index. Three junior pharmacists independently reviewed each vignette under simulated conditions. Main Outcomes and Measures: The study assesses the LLM-based CDSS's accuracy, precision, recall, and F1 scores in identifying Drug-Related Problems (DRPs), compared to junior pharmacists alone or in an assistive mode with the CDSS. Results: The co-pilot mode of RAG-LLM significantly improved DRP identification accuracy by 22% over solo pharmacists. It showed higher recall and F1 scores, indicating better detection of severe DRPs, despite a slight decrease in precision. Accuracy varied across categories when pharmacists had access to RAG-LLM responses. Conclusions: The RAG-LLM based CDSS enhances medication error identification accuracy when used with junior pharmacists, especially in detecting severe DRPs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge