Nigam Shah
TIMER: Temporal Instruction Modeling and Evaluation for Longitudinal Clinical Records
Mar 06, 2025Abstract:Large language models (LLMs) have emerged as promising tools for assisting in medical tasks, yet processing Electronic Health Records (EHRs) presents unique challenges due to their longitudinal nature. While LLMs' capabilities to perform medical tasks continue to improve, their ability to reason over temporal dependencies across multiple patient visits and time frames remains unexplored. We introduce TIMER (Temporal Instruction Modeling and Evaluation for Longitudinal Clinical Records), a framework that incorporate instruction-response pairs grounding to different parts of a patient's record as a critical dimension in both instruction evaluation and tuning for longitudinal clinical records. We develop TIMER-Bench, the first time-aware benchmark that evaluates temporal reasoning capabilities over longitudinal EHRs, as well as TIMER-Instruct, an instruction-tuning methodology for LLMs to learn reasoning over time. We demonstrate that models fine-tuned with TIMER-Instruct improve performance by 7.3% on human-generated benchmarks and 9.2% on TIMER-Bench, indicating that temporal instruction-tuning improves model performance for reasoning over EHR.
VeriFact: Verifying Facts in LLM-Generated Clinical Text with Electronic Health Records
Jan 28, 2025Abstract:Methods to ensure factual accuracy of text generated by large language models (LLM) in clinical medicine are lacking. VeriFact is an artificial intelligence system that combines retrieval-augmented generation and LLM-as-a-Judge to verify whether LLM-generated text is factually supported by a patient's medical history based on their electronic health record (EHR). To evaluate this system, we introduce VeriFact-BHC, a new dataset that decomposes Brief Hospital Course narratives from discharge summaries into a set of simple statements with clinician annotations for whether each statement is supported by the patient's EHR clinical notes. Whereas highest agreement between clinicians was 88.5%, VeriFact achieves up to 92.7% agreement when compared to a denoised and adjudicated average human clinican ground truth, suggesting that VeriFact exceeds the average clinician's ability to fact-check text against a patient's medical record. VeriFact may accelerate the development of LLM-based EHR applications by removing current evaluation bottlenecks.
Assessing the Limitations of Large Language Models in Clinical Fact Decomposition
Dec 17, 2024Abstract:Verifying factual claims is critical for using large language models (LLMs) in healthcare. Recent work has proposed fact decomposition, which uses LLMs to rewrite source text into concise sentences conveying a single piece of information, as an approach for fine-grained fact verification. Clinical documentation poses unique challenges for fact decomposition due to dense terminology and diverse note types. To explore these challenges, we present FactEHR, a dataset consisting of full document fact decompositions for 2,168 clinical notes spanning four types from three hospital systems. Our evaluation, including review by clinicians, highlights significant variability in the quality of fact decomposition for four commonly used LLMs, with some LLMs generating 2.6x more facts per sentence than others. The results underscore the need for better LLM capabilities to support factual verification in clinical text. To facilitate future research in this direction, we plan to release our code at \url{https://github.com/som-shahlab/factehr}.
A Proposed S.C.O.R.E. Evaluation Framework for Large Language Models : Safety, Consensus, Objectivity, Reproducibility and Explainability
Jul 10, 2024



Abstract:A comprehensive qualitative evaluation framework for large language models (LLM) in healthcare that expands beyond traditional accuracy and quantitative metrics needed. We propose 5 key aspects for evaluation of LLMs: Safety, Consensus, Objectivity, Reproducibility and Explainability (S.C.O.R.E.). We suggest that S.C.O.R.E. may form the basis for an evaluation framework for future LLM-based models that are safe, reliable, trustworthy, and ethical for healthcare and clinical applications.
A Multi-Center Study on the Adaptability of a Shared Foundation Model for Electronic Health Records
Nov 20, 2023Abstract:Foundation models hold promise for transforming AI in healthcare by providing modular components that are easily adaptable to downstream healthcare tasks, making AI development more scalable and cost-effective. Structured EHR foundation models, trained on coded medical records from millions of patients, demonstrated benefits including increased performance with fewer training labels, and improved robustness to distribution shifts. However, questions remain on the feasibility of sharing these models across different hospitals and their performance for local task adaptation. This multi-center study examined the adaptability of a recently released structured EHR foundation model ($FM_{SM}$), trained on longitudinal medical record data from 2.57M Stanford Medicine patients. Experiments were conducted using EHR data at The Hospital for Sick Children and MIMIC-IV. We assessed both adaptability via continued pretraining on local data, and task adaptability compared to baselines of training models from scratch at each site, including a local foundation model. We evaluated the performance of these models on 8 clinical prediction tasks. In both datasets, adapting the off-the-shelf $FM_{SM}$ matched the performance of GBM models locally trained on all data while providing a 13% improvement in settings with few task-specific training labels. With continued pretraining on local data, label efficiency substantially improved, such that $FM_{SM}$ required fewer than 1% of training examples to match the fully trained GBM's performance. Continued pretraining was also 60 to 90% more sample-efficient than training local foundation models from scratch. Our findings show that adapting shared EHR foundation models across hospitals provides improved prediction performance at less cost, underscoring the utility of base foundation models as modular components to streamline the development of healthcare AI.
Clinfo.ai: An Open-Source Retrieval-Augmented Large Language Model System for Answering Medical Questions using Scientific Literature
Oct 24, 2023



Abstract:The quickly-expanding nature of published medical literature makes it challenging for clinicians and researchers to keep up with and summarize recent, relevant findings in a timely manner. While several closed-source summarization tools based on large language models (LLMs) now exist, rigorous and systematic evaluations of their outputs are lacking. Furthermore, there is a paucity of high-quality datasets and appropriate benchmark tasks with which to evaluate these tools. We address these issues with four contributions: we release Clinfo.ai, an open-source WebApp that answers clinical questions based on dynamically retrieved scientific literature; we specify an information retrieval and abstractive summarization task to evaluate the performance of such retrieval-augmented LLM systems; we release a dataset of 200 questions and corresponding answers derived from published systematic reviews, which we name PubMed Retrieval and Synthesis (PubMedRS-200); and report benchmark results for Clinfo.ai and other publicly available OpenQA systems on PubMedRS-200.
EHRSHOT: An EHR Benchmark for Few-Shot Evaluation of Foundation Models
Jul 05, 2023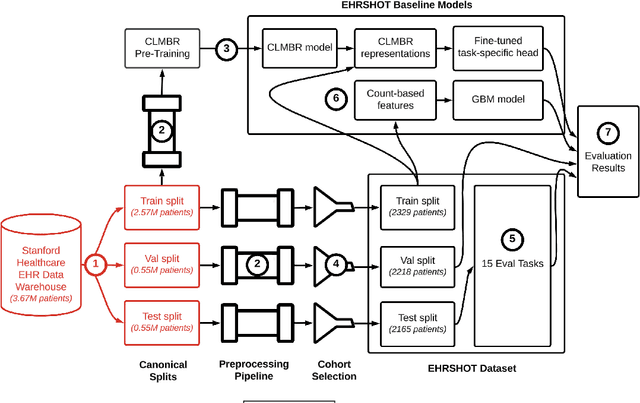
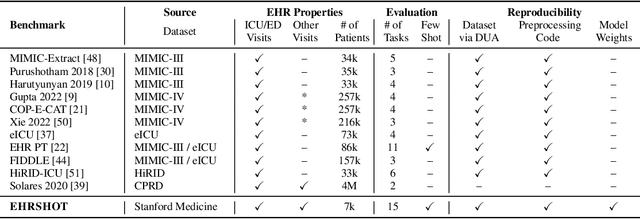

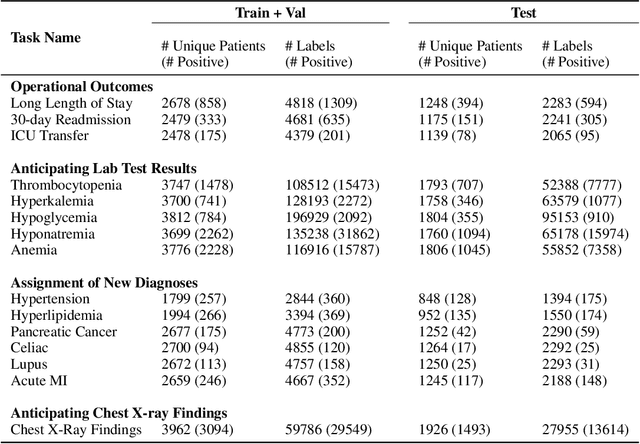
Abstract:While the general machine learning (ML) community has benefited from public datasets, tasks, and models, the progress of ML in healthcare has been hampered by a lack of such shared assets. The success of foundation models creates new challenges for healthcare ML by requiring access to shared pretrained models to validate performance benefits. We help address these challenges through three contributions. First, we publish a new dataset, EHRSHOT, containing de-identified structured data from the electronic health records (EHRs) of 6,712 patients from Stanford Medicine. Unlike MIMIC-III/IV and other popular EHR datasets, EHRSHOT is longitudinal and not restricted to ICU/ED patients. Second, we publish the weights of a 141M parameter clinical foundation model pretrained on the structured EHR data of 2.57M patients. We are one of the first to fully release such a model for coded EHR data; in contrast, most prior models released for clinical data (e.g. GatorTron, ClinicalBERT) only work with unstructured text and cannot process the rich, structured data within an EHR. We provide an end-to-end pipeline for the community to validate and build upon its performance. Third, we define 15 few-shot clinical prediction tasks, enabling evaluation of foundation models on benefits such as sample efficiency and task adaption. The code to reproduce our results, as well as the model and dataset (via a research data use agreement), are available at our Github repo here: https://github.com/som-shahlab/ehrshot-benchmark
Self-Supervised Time-to-Event Modeling with Structured Medical Records
Jan 09, 2023



Abstract:Time-to-event models (also known as survival models) are used in medicine and other fields for estimating the probability distribution of the time until a particular event occurs. While providing many advantages over traditional classification models, such as naturally handling censoring, time-to-event models require more parameters and are challenging to learn in settings with limited labeled training data. High censoring rates, common in events with long time horizons, further limit available training data and exacerbate the risk of overfitting. Existing methods, such as proportional hazard or accelerated failure time-based approaches, employ distributional assumptions to reduce parameter size, but they are vulnerable to model misspecification. In this work, we address these challenges with MOTOR, a self-supervised model that leverages temporal structure found in large-scale collections of timestamped, but largely unlabeled events, typical of electronic health record data. MOTOR defines a time-to-event pretraining task that naturally captures the probability distribution of event times, making it well-suited to applications in medicine. After pretraining on 8,192 tasks auto-generated from 2.7M patients (2.4B clinical events), we evaluate the performance of our pretrained model after fine-tuning to unseen time-to-event tasks. MOTOR-derived models improve upon current state-of-the-art C statistic performance by 6.6% and decrease training time (in wall time) by up to 8.2 times. We further improve sample efficiency, with adapted models matching current state-of-the-art performance using 95% less training data.
RadFusion: Benchmarking Performance and Fairness for Multimodal Pulmonary Embolism Detection from CT and EHR
Nov 27, 2021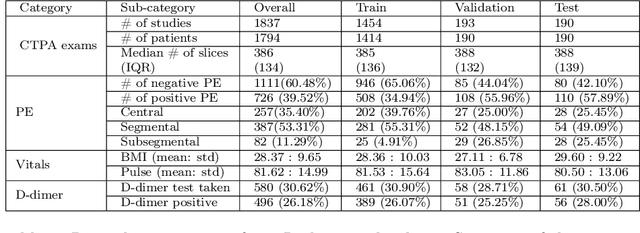
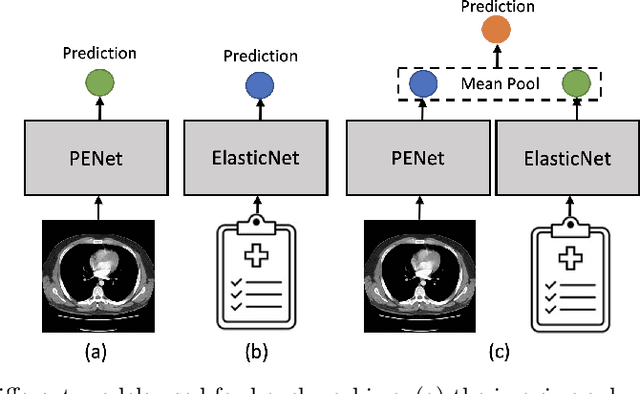
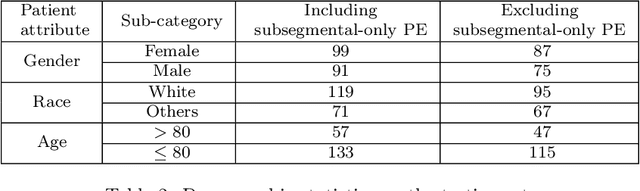
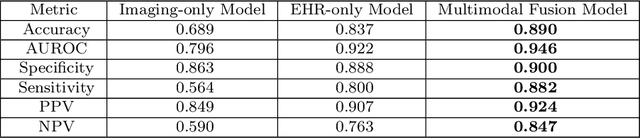
Abstract:Despite the routine use of electronic health record (EHR) data by radiologists to contextualize clinical history and inform image interpretation, the majority of deep learning architectures for medical imaging are unimodal, i.e., they only learn features from pixel-level information. Recent research revealing how race can be recovered from pixel data alone highlights the potential for serious biases in models which fail to account for demographics and other key patient attributes. Yet the lack of imaging datasets which capture clinical context, inclusive of demographics and longitudinal medical history, has left multimodal medical imaging underexplored. To better assess these challenges, we present RadFusion, a multimodal, benchmark dataset of 1794 patients with corresponding EHR data and high-resolution computed tomography (CT) scans labeled for pulmonary embolism. We evaluate several representative multimodal fusion models and benchmark their fairness properties across protected subgroups, e.g., gender, race/ethnicity, age. Our results suggest that integrating imaging and EHR data can improve classification performance and robustness without introducing large disparities in the true positive rate between population groups.
Evaluating Treatment Prioritization Rules via Rank-Weighted Average Treatment Effects
Nov 15, 2021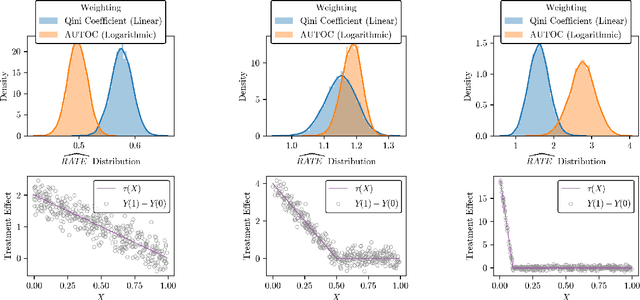
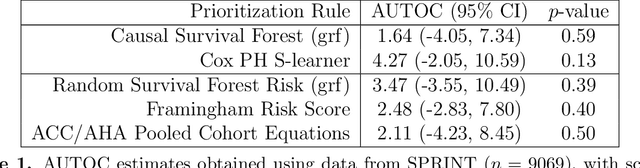
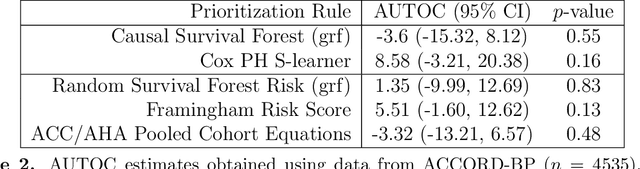
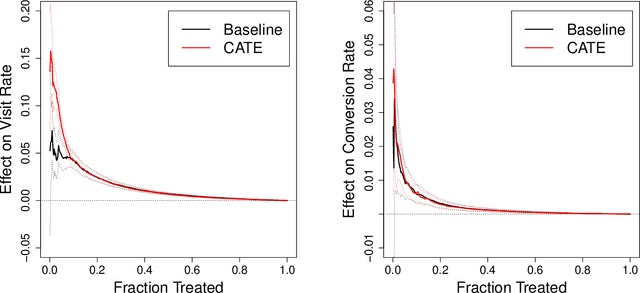
Abstract:There are a number of available methods that can be used for choosing whom to prioritize treatment, including ones based on treatment effect estimation, risk scoring, and hand-crafted rules. We propose rank-weighted average treatment effect (RATE) metrics as a simple and general family of metrics for comparing treatment prioritization rules on a level playing field. RATEs are agnostic as to how the prioritization rules were derived, and only assesses them based on how well they succeed in identifying units that benefit the most from treatment. We define a family of RATE estimators and prove a central limit theorem that enables asymptotically exact inference in a wide variety of randomized and observational study settings. We provide justification for the use of bootstrapped confidence intervals and a framework for testing hypotheses about heterogeneity in treatment effectiveness correlated with the prioritization rule. Our definition of the RATE nests a number of existing metrics, including the Qini coefficient, and our analysis directly yields inference methods for these metrics. We demonstrate our approach in examples drawn from both personalized medicine and marketing. In the medical setting, using data from the SPRINT and ACCORD-BP randomized control trials, we find no significant evidence of heterogeneous treatment effects. On the other hand, in a large marketing trial, we find robust evidence of heterogeneity in the treatment effects of some digital advertising campaigns and demonstrate how RATEs can be used to compare targeting rules that prioritize estimated risk vs. those that prioritize estimated treatment benefit.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge