Alejandro Lozano
PaperSearchQA: Learning to Search and Reason over Scientific Papers with RLVR
Jan 26, 2026Abstract:Search agents are language models (LMs) that reason and search knowledge bases (or the web) to answer questions; recent methods supervise only the final answer accuracy using reinforcement learning with verifiable rewards (RLVR). Most RLVR search agents tackle general-domain QA, which limits their relevance to technical AI systems in science, engineering, and medicine. In this work we propose training agents to search and reason over scientific papers -- this tests technical question-answering, it is directly relevant to real scientists, and the capabilities will be crucial to future AI Scientist systems. Concretely, we release a search corpus of 16 million biomedical paper abstracts and construct a challenging factoid QA dataset called PaperSearchQA with 60k samples answerable from the corpus, along with benchmarks. We train search agents in this environment to outperform non-RL retrieval baselines; we also perform further quantitative analysis and observe interesting agent behaviors like planning, reasoning, and self-verification. Our corpus, datasets, and benchmarks are usable with the popular Search-R1 codebase for RLVR training and released on https://huggingface.co/collections/jmhb/papersearchqa. Finally, our data creation methods are scalable and easily extendable to other scientific domains.
Can Large Language Models Match the Conclusions of Systematic Reviews?
May 28, 2025Abstract:Systematic reviews (SR), in which experts summarize and analyze evidence across individual studies to provide insights on a specialized topic, are a cornerstone for evidence-based clinical decision-making, research, and policy. Given the exponential growth of scientific articles, there is growing interest in using large language models (LLMs) to automate SR generation. However, the ability of LLMs to critically assess evidence and reason across multiple documents to provide recommendations at the same proficiency as domain experts remains poorly characterized. We therefore ask: Can LLMs match the conclusions of systematic reviews written by clinical experts when given access to the same studies? To explore this question, we present MedEvidence, a benchmark pairing findings from 100 SRs with the studies they are based on. We benchmark 24 LLMs on MedEvidence, including reasoning, non-reasoning, medical specialist, and models across varying sizes (from 7B-700B). Through our systematic evaluation, we find that reasoning does not necessarily improve performance, larger models do not consistently yield greater gains, and knowledge-based fine-tuning degrades accuracy on MedEvidence. Instead, most models exhibit similar behavior: performance tends to degrade as token length increases, their responses show overconfidence, and, contrary to human experts, all models show a lack of scientific skepticism toward low-quality findings. These results suggest that more work is still required before LLMs can reliably match the observations from expert-conducted SRs, even though these systems are already deployed and being used by clinicians. We release our codebase and benchmark to the broader research community to further investigate LLM-based SR systems.
MedCaseReasoning: Evaluating and learning diagnostic reasoning from clinical case reports
May 16, 2025Abstract:Doctors and patients alike increasingly use Large Language Models (LLMs) to diagnose clinical cases. However, unlike domains such as math or coding, where correctness can be objectively defined by the final answer, medical diagnosis requires both the outcome and the reasoning process to be accurate. Currently, widely used medical benchmarks like MedQA and MMLU assess only accuracy in the final answer, overlooking the quality and faithfulness of the clinical reasoning process. To address this limitation, we introduce MedCaseReasoning, the first open-access dataset for evaluating LLMs on their ability to align with clinician-authored diagnostic reasoning. The dataset includes 14,489 diagnostic question-and-answer cases, each paired with detailed reasoning statements derived from open-access medical case reports. We evaluate state-of-the-art reasoning LLMs on MedCaseReasoning and find significant shortcomings in their diagnoses and reasoning: for instance, the top-performing open-source model, DeepSeek-R1, achieves only 48% 10-shot diagnostic accuracy and mentions only 64% of the clinician reasoning statements (recall). However, we demonstrate that fine-tuning LLMs on the reasoning traces derived from MedCaseReasoning significantly improves diagnostic accuracy and clinical reasoning recall by an average relative gain of 29% and 41%, respectively. The open-source dataset, code, and models are available at https://github.com/kevinwu23/Stanford-MedCaseReasoning.
MicroVQA: A Multimodal Reasoning Benchmark for Microscopy-Based Scientific Research
Mar 17, 2025Abstract:Scientific research demands sophisticated reasoning over multimodal data, a challenge especially prevalent in biology. Despite recent advances in multimodal large language models (MLLMs) for AI-assisted research, existing multimodal reasoning benchmarks only target up to college-level difficulty, while research-level benchmarks emphasize lower-level perception, falling short of the complex multimodal reasoning needed for scientific discovery. To bridge this gap, we introduce MicroVQA, a visual-question answering (VQA) benchmark designed to assess three reasoning capabilities vital in research workflows: expert image understanding, hypothesis generation, and experiment proposal. MicroVQA consists of 1,042 multiple-choice questions (MCQs) curated by biology experts across diverse microscopy modalities, ensuring VQA samples represent real scientific practice. In constructing the benchmark, we find that standard MCQ generation methods induce language shortcuts, motivating a new two-stage pipeline: an optimized LLM prompt structures question-answer pairs into MCQs; then, an agent-based `RefineBot' updates them to remove shortcuts. Benchmarking on state-of-the-art MLLMs reveal a peak performance of 53\%; models with smaller LLMs only slightly underperform top models, suggesting that language-based reasoning is less challenging than multimodal reasoning; and tuning with scientific articles enhances performance. Expert analysis of chain-of-thought responses shows that perception errors are the most frequent, followed by knowledge errors and then overgeneralization errors. These insights highlight the challenges in multimodal scientific reasoning, showing MicroVQA is a valuable resource advancing AI-driven biomedical research. MicroVQA is available at https://huggingface.co/datasets/jmhb/microvqa, and project page at https://jmhb0.github.io/microvqa.
Video Action Differencing
Mar 10, 2025Abstract:How do two individuals differ when performing the same action? In this work, we introduce Video Action Differencing (VidDiff), the novel task of identifying subtle differences between videos of the same action, which has many applications, such as coaching and skill learning. To enable development on this new task, we first create VidDiffBench, a benchmark dataset containing 549 video pairs, with human annotations of 4,469 fine-grained action differences and 2,075 localization timestamps indicating where these differences occur. Our experiments demonstrate that VidDiffBench poses a significant challenge for state-of-the-art large multimodal models (LMMs), such as GPT-4o and Qwen2-VL. By analyzing failure cases of LMMs on VidDiffBench, we highlight two key challenges for this task: localizing relevant sub-actions over two videos and fine-grained frame comparison. To overcome these, we propose the VidDiff method, an agentic workflow that breaks the task into three stages: action difference proposal, keyframe localization, and frame differencing, each stage utilizing specialized foundation models. To encourage future research in this new task, we release the benchmark at https://huggingface.co/datasets/jmhb/VidDiffBench and code at http://jmhb0.github.io/viddiff.
BIOMEDICA: An Open Biomedical Image-Caption Archive, Dataset, and Vision-Language Models Derived from Scientific Literature
Jan 14, 2025Abstract:The development of vision-language models (VLMs) is driven by large-scale and diverse multimodal datasets. However, progress toward generalist biomedical VLMs is limited by the lack of annotated, publicly accessible datasets across biology and medicine. Existing efforts are restricted to narrow domains, missing the full diversity of biomedical knowledge encoded in scientific literature. To address this gap, we introduce BIOMEDICA, a scalable, open-source framework to extract, annotate, and serialize the entirety of the PubMed Central Open Access subset into an easy-to-use, publicly accessible dataset. Our framework produces a comprehensive archive with over 24 million unique image-text pairs from over 6 million articles. Metadata and expert-guided annotations are also provided. We demonstrate the utility and accessibility of our resource by releasing BMCA-CLIP, a suite of CLIP-style models continuously pre-trained on the BIOMEDICA dataset via streaming, eliminating the need to download 27 TB of data locally. On average, our models achieve state-of-the-art performance across 40 tasks - spanning pathology, radiology, ophthalmology, dermatology, surgery, molecular biology, parasitology, and cell biology - excelling in zero-shot classification with a 6.56% average improvement (as high as 29.8% and 17.5% in dermatology and ophthalmology, respectively), and stronger image-text retrieval, all while using 10x less compute. To foster reproducibility and collaboration, we release our codebase and dataset for the broader research community.
Automated Generation of Challenging Multiple-Choice Questions for Vision Language Model Evaluation
Jan 06, 2025



Abstract:The rapid development of vision language models (VLMs) demands rigorous and reliable evaluation. However, current visual question answering (VQA) benchmarks often depend on open-ended questions, making accurate evaluation difficult due to the variability in natural language responses. To address this, we introduce AutoConverter, an agentic framework that automatically converts these open-ended questions into multiple-choice format, enabling objective evaluation while reducing the costly question creation process. Our experiments demonstrate that AutoConverter can generate correct and challenging multiple-choice questions, with VLMs demonstrating consistently similar or lower accuracy on these questions compared to human-created ones. Using AutoConverter, we construct VMCBench, a benchmark created by transforming 20 existing VQA datasets into a unified multiple-choice format, totaling 9,018 questions. We comprehensively evaluate 33 state-of-the-art VLMs on VMCBench, setting a new standard for scalable, consistent, and reproducible VLM evaluation.
Time-to-Event Pretraining for 3D Medical Imaging
Nov 14, 2024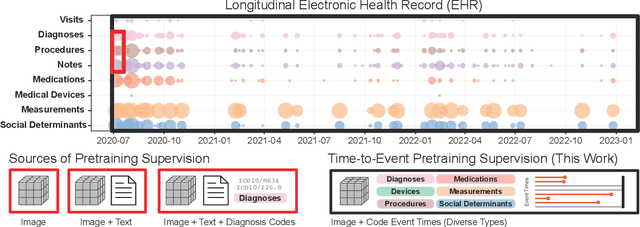
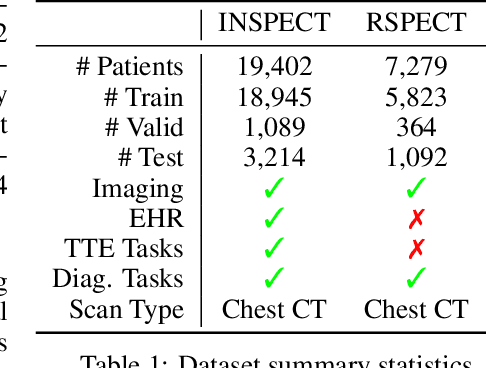
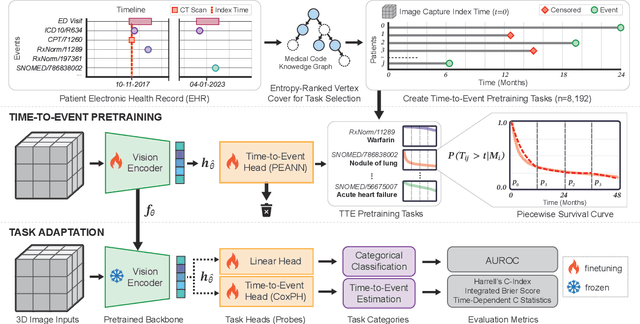
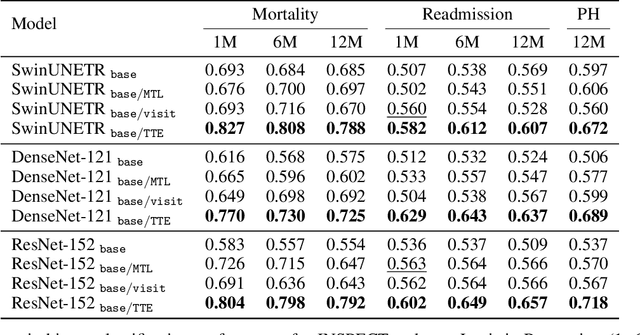
Abstract:With the rise of medical foundation models and the growing availability of imaging data, scalable pretraining techniques offer a promising way to identify imaging biomarkers predictive of future disease risk. While current self-supervised methods for 3D medical imaging models capture local structural features like organ morphology, they fail to link pixel biomarkers with long-term health outcomes due to a missing context problem. Current approaches lack the temporal context necessary to identify biomarkers correlated with disease progression, as they rely on supervision derived only from images and concurrent text descriptions. To address this, we introduce time-to-event pretraining, a pretraining framework for 3D medical imaging models that leverages large-scale temporal supervision from paired, longitudinal electronic health records (EHRs). Using a dataset of 18,945 CT scans (4.2 million 2D images) and time-to-event distributions across thousands of EHR-derived tasks, our method improves outcome prediction, achieving an average AUROC increase of 23.7% and a 29.4% gain in Harrell's C-index across 8 benchmark tasks. Importantly, these gains are achieved without sacrificing diagnostic classification performance. This study lays the foundation for integrating longitudinal EHR and 3D imaging data to advance clinical risk prediction.
μ-Bench: A Vision-Language Benchmark for Microscopy Understanding
Jul 01, 2024



Abstract:Recent advances in microscopy have enabled the rapid generation of terabytes of image data in cell biology and biomedical research. Vision-language models (VLMs) offer a promising solution for large-scale biological image analysis, enhancing researchers' efficiency, identifying new image biomarkers, and accelerating hypothesis generation and scientific discovery. However, there is a lack of standardized, diverse, and large-scale vision-language benchmarks to evaluate VLMs' perception and cognition capabilities in biological image understanding. To address this gap, we introduce {\mu}-Bench, an expert-curated benchmark encompassing 22 biomedical tasks across various scientific disciplines (biology, pathology), microscopy modalities (electron, fluorescence, light), scales (subcellular, cellular, tissue), and organisms in both normal and abnormal states. We evaluate state-of-the-art biomedical, pathology, and general VLMs on {\mu}-Bench and find that: i) current models struggle on all categories, even for basic tasks such as distinguishing microscopy modalities; ii) current specialist models fine-tuned on biomedical data often perform worse than generalist models; iii) fine-tuning in specific microscopy domains can cause catastrophic forgetting, eroding prior biomedical knowledge encoded in their base model. iv) weight interpolation between fine-tuned and pre-trained models offers one solution to forgetting and improves general performance across biomedical tasks. We release {\mu}-Bench under a permissive license to accelerate the research and development of microscopy foundation models.
Recent Advances, Applications, and Open Challenges in Machine Learning for Health: Reflections from Research Roundtables at ML4H 2023 Symposium
Mar 03, 2024Abstract:The third ML4H symposium was held in person on December 10, 2023, in New Orleans, Louisiana, USA. The symposium included research roundtable sessions to foster discussions between participants and senior researchers on timely and relevant topics for the \ac{ML4H} community. Encouraged by the successful virtual roundtables in the previous year, we organized eleven in-person roundtables and four virtual roundtables at ML4H 2022. The organization of the research roundtables at the conference involved 17 Senior Chairs and 19 Junior Chairs across 11 tables. Each roundtable session included invited senior chairs (with substantial experience in the field), junior chairs (responsible for facilitating the discussion), and attendees from diverse backgrounds with interest in the session's topic. Herein we detail the organization process and compile takeaways from these roundtable discussions, including recent advances, applications, and open challenges for each topic. We conclude with a summary and lessons learned across all roundtables. This document serves as a comprehensive review paper, summarizing the recent advancements in machine learning for healthcare as contributed by foremost researchers in the field.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge