Kenneth W. Mahaffey
Zero-Shot Clinical Trial Patient Matching with LLMs
Feb 05, 2024
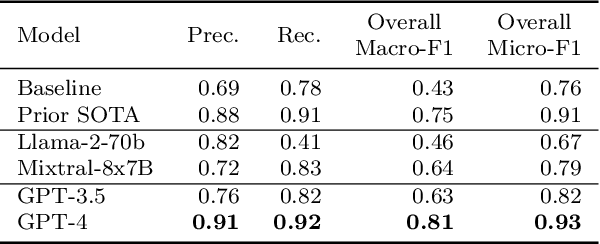
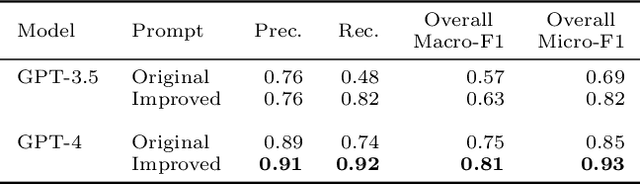

Abstract:Matching patients to clinical trials is a key unsolved challenge in bringing new drugs to market. Today, identifying patients who meet a trial's eligibility criteria is highly manual, taking up to 1 hour per patient. Automated screening is challenging, however, as it requires understanding unstructured clinical text. Large language models (LLMs) offer a promising solution. In this work, we explore their application to trial matching. First, we design an LLM-based system which, given a patient's medical history as unstructured clinical text, evaluates whether that patient meets a set of inclusion criteria (also specified as free text). Our zero-shot system achieves state-of-the-art scores on the n2c2 2018 cohort selection benchmark. Second, we improve the data and cost efficiency of our method by identifying a prompting strategy which matches patients an order of magnitude faster and more cheaply than the status quo, and develop a two-stage retrieval pipeline that reduces the number of tokens processed by up to a third while retaining high performance. Third, we evaluate the interpretability of our system by having clinicians evaluate the natural language justifications generated by the LLM for each eligibility decision, and show that it can output coherent explanations for 97% of its correct decisions and 75% of its incorrect ones. Our results establish the feasibility of using LLMs to accelerate clinical trial operations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge