Michael Wornow
Cost-Efficient Serving of LLM Agents via Test-Time Plan Caching
Jun 17, 2025Abstract:LLM-based agentic applications have shown increasingly remarkable capabilities in complex workflows but incur substantial costs due to extensive planning and reasoning requirements. Existing LLM caching techniques (like context caching and semantic caching), primarily designed for serving chatbots, are insufficient for agentic applications where outputs depend on external data or environmental contexts. We propose agentic plan caching, a novel approach that extracts, stores, adapts, and reuses structured plan templates from planning stages of agentic applications across semantically similar tasks to reduce the cost of serving. Unlike traditional semantic caching, our system extracts plan templates from completed agent executions at test-time, employs keyword extraction to match new requests against cached plans, and utilizes lightweight models to adapt these templates to task-specific plans with contexts. Evaluation across multiple real-world agentic applications shows that our system can reduce costs by 46.62% on average while maintaining performance, offering a more efficient solution for serving LLM-based agents that complements existing LLM serving infrastructures.
MedHELM: Holistic Evaluation of Large Language Models for Medical Tasks
May 26, 2025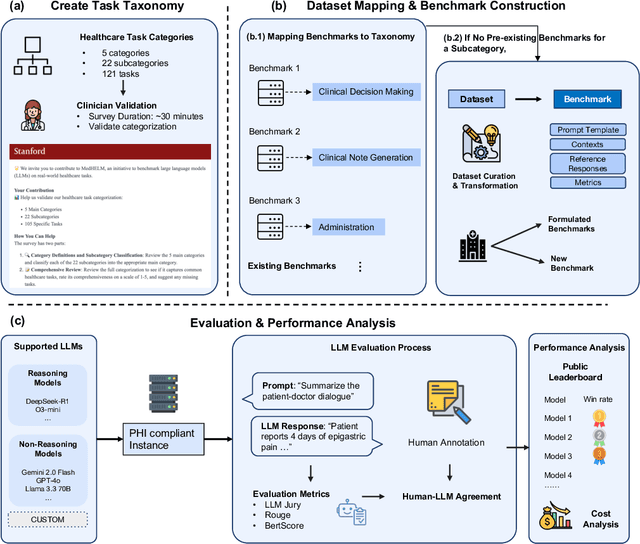
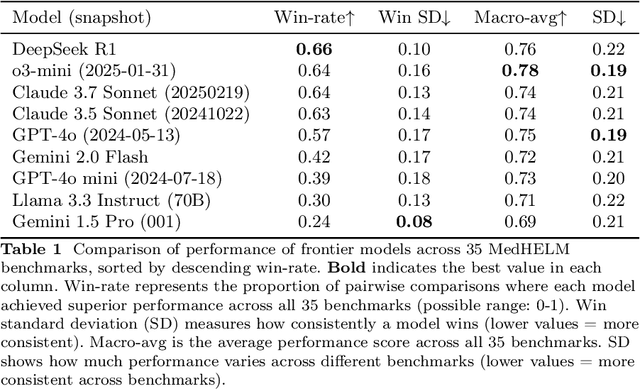
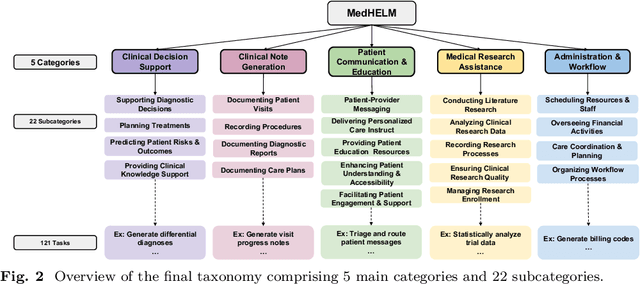
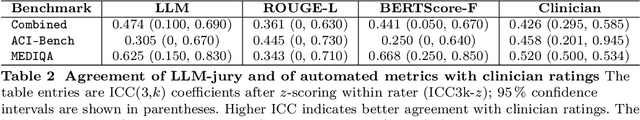
Abstract:While large language models (LLMs) achieve near-perfect scores on medical licensing exams, these evaluations inadequately reflect the complexity and diversity of real-world clinical practice. We introduce MedHELM, an extensible evaluation framework for assessing LLM performance for medical tasks with three key contributions. First, a clinician-validated taxonomy spanning 5 categories, 22 subcategories, and 121 tasks developed with 29 clinicians. Second, a comprehensive benchmark suite comprising 35 benchmarks (17 existing, 18 newly formulated) providing complete coverage of all categories and subcategories in the taxonomy. Third, a systematic comparison of LLMs with improved evaluation methods (using an LLM-jury) and a cost-performance analysis. Evaluation of 9 frontier LLMs, using the 35 benchmarks, revealed significant performance variation. Advanced reasoning models (DeepSeek R1: 66% win-rate; o3-mini: 64% win-rate) demonstrated superior performance, though Claude 3.5 Sonnet achieved comparable results at 40% lower estimated computational cost. On a normalized accuracy scale (0-1), most models performed strongly in Clinical Note Generation (0.73-0.85) and Patient Communication & Education (0.78-0.83), moderately in Medical Research Assistance (0.65-0.75), and generally lower in Clinical Decision Support (0.56-0.72) and Administration & Workflow (0.53-0.63). Our LLM-jury evaluation method achieved good agreement with clinician ratings (ICC = 0.47), surpassing both average clinician-clinician agreement (ICC = 0.43) and automated baselines including ROUGE-L (0.36) and BERTScore-F1 (0.44). Claude 3.5 Sonnet achieved comparable performance to top models at lower estimated cost. These findings highlight the importance of real-world, task-specific evaluation for medical use of LLMs and provides an open source framework to enable this.
Context Clues: Evaluating Long Context Models for Clinical Prediction Tasks on EHRs
Dec 09, 2024



Abstract:Foundation Models (FMs) trained on Electronic Health Records (EHRs) have achieved state-of-the-art results on numerous clinical prediction tasks. However, most existing EHR FMs have context windows of <1k tokens. This prevents them from modeling full patient EHRs which can exceed 10k's of events. Recent advancements in subquadratic long-context architectures (e.g., Mamba) offer a promising solution. However, their application to EHR data has not been well-studied. We address this gap by presenting the first systematic evaluation of the effect of context length on modeling EHR data. We find that longer context models improve predictive performance -- our Mamba-based model surpasses the prior state-of-the-art on 9/14 tasks on the EHRSHOT prediction benchmark. For clinical applications, however, model performance alone is insufficient -- robustness to the unique properties of EHR is crucial. Thus, we also evaluate models across three previously underexplored properties of EHR data: (1) the prevalence of "copy-forwarded" diagnoses which creates artificial repetition of tokens within EHR sequences; (2) the irregular time intervals between EHR events which can lead to a wide range of timespans within a context window; and (3) the natural increase in disease complexity over time which makes later tokens in the EHR harder to predict than earlier ones. Stratifying our EHRSHOT results, we find that higher levels of each property correlate negatively with model performance, but that longer context models are more robust to more extreme levels of these properties. Our work highlights the potential for using long-context architectures to model EHR data, and offers a case study for identifying new challenges in modeling sequential data motivated by domains outside of natural language. We release our models and code at: https://github.com/som-shahlab/long_context_clues
meds_reader: A fast and efficient EHR processing library
Sep 12, 2024



Abstract:The growing demand for machine learning in healthcare requires processing increasingly large electronic health record (EHR) datasets, but existing pipelines are not computationally efficient or scalable. In this paper, we introduce meds_reader, an optimized Python package for efficient EHR data processing that is designed to take advantage of many intrinsic properties of EHR data for improved speed. We then demonstrate the benefits of meds_reader by reimplementing key components of two major EHR processing pipelines, achieving 10-100x improvements in memory, speed, and disk usage. The code for meds_reader can be found at https://github.com/som-shahlab/meds_reader.
Do Multimodal Foundation Models Understand Enterprise Workflows? A Benchmark for Business Process Management Tasks
Jun 19, 2024



Abstract:Existing ML benchmarks lack the depth and diversity of annotations needed for evaluating models on business process management (BPM) tasks. BPM is the practice of documenting, measuring, improving, and automating enterprise workflows. However, research has focused almost exclusively on one task - full end-to-end automation using agents based on multimodal foundation models (FMs) like GPT-4. This focus on automation ignores the reality of how most BPM tools are applied today - simply documenting the relevant workflow takes 60% of the time of the typical process optimization project. To address this gap we present WONDERBREAD, the first benchmark for evaluating multimodal FMs on BPM tasks beyond automation. Our contributions are: (1) a dataset containing 2928 documented workflow demonstrations; (2) 6 novel BPM tasks sourced from real-world applications ranging from workflow documentation to knowledge transfer to process improvement; and (3) an automated evaluation harness. Our benchmark shows that while state-of-the-art FMs can automatically generate documentation (e.g. recalling 88% of the steps taken in a video demonstration of a workflow), they struggle to re-apply that knowledge towards finer-grained validation of workflow completion (F1 < 0.3). We hope WONDERBREAD encourages the development of more "human-centered" AI tooling for enterprise applications and furthers the exploration of multimodal FMs for the broader universe of BPM tasks. We publish our dataset and experiments here: https://github.com/HazyResearch/wonderbread
Automating the Enterprise with Foundation Models
May 03, 2024



Abstract:Automating enterprise workflows could unlock $4 trillion/year in productivity gains. Despite being of interest to the data management community for decades, the ultimate vision of end-to-end workflow automation has remained elusive. Current solutions rely on process mining and robotic process automation (RPA), in which a bot is hard-coded to follow a set of predefined rules for completing a workflow. Through case studies of a hospital and large B2B enterprise, we find that the adoption of RPA has been inhibited by high set-up costs (12-18 months), unreliable execution (60% initial accuracy), and burdensome maintenance (requiring multiple FTEs). Multimodal foundation models (FMs) such as GPT-4 offer a promising new approach for end-to-end workflow automation given their generalized reasoning and planning abilities. To study these capabilities we propose ECLAIR, a system to automate enterprise workflows with minimal human supervision. We conduct initial experiments showing that multimodal FMs can address the limitations of traditional RPA with (1) near-human-level understanding of workflows (93% accuracy on a workflow understanding task) and (2) instant set-up with minimal technical barrier (based solely on a natural language description of a workflow, ECLAIR achieves end-to-end completion rates of 40%). We identify human-AI collaboration, validation, and self-improvement as open challenges, and suggest ways they can be solved with data management techniques. Code is available at: https://github.com/HazyResearch/eclair-agents
Standing on FURM ground -- A framework for evaluating Fair, Useful, and Reliable AI Models in healthcare systems
Mar 14, 2024



Abstract:The impact of using artificial intelligence (AI) to guide patient care or operational processes is an interplay of the AI model's output, the decision-making protocol based on that output, and the capacity of the stakeholders involved to take the necessary subsequent action. Estimating the effects of this interplay before deployment, and studying it in real time afterwards, are essential to bridge the chasm between AI model development and achievable benefit. To accomplish this, the Data Science team at Stanford Health Care has developed a Testing and Evaluation (T&E) mechanism to identify fair, useful and reliable AI models (FURM) by conducting an ethical review to identify potential value mismatches, simulations to estimate usefulness, financial projections to assess sustainability, as well as analyses to determine IT feasibility, design a deployment strategy, and recommend a prospective monitoring and evaluation plan. We report on FURM assessments done to evaluate six AI guided solutions for potential adoption, spanning clinical and operational settings, each with the potential to impact from several dozen to tens of thousands of patients each year. We describe the assessment process, summarize the six assessments, and share our framework to enable others to conduct similar assessments. Of the six solutions we assessed, two have moved into a planning and implementation phase. Our novel contributions - usefulness estimates by simulation, financial projections to quantify sustainability, and a process to do ethical assessments - as well as their underlying methods and open source tools, are available for other healthcare systems to conduct actionable evaluations of candidate AI solutions.
Recent Advances, Applications, and Open Challenges in Machine Learning for Health: Reflections from Research Roundtables at ML4H 2023 Symposium
Mar 03, 2024Abstract:The third ML4H symposium was held in person on December 10, 2023, in New Orleans, Louisiana, USA. The symposium included research roundtable sessions to foster discussions between participants and senior researchers on timely and relevant topics for the \ac{ML4H} community. Encouraged by the successful virtual roundtables in the previous year, we organized eleven in-person roundtables and four virtual roundtables at ML4H 2022. The organization of the research roundtables at the conference involved 17 Senior Chairs and 19 Junior Chairs across 11 tables. Each roundtable session included invited senior chairs (with substantial experience in the field), junior chairs (responsible for facilitating the discussion), and attendees from diverse backgrounds with interest in the session's topic. Herein we detail the organization process and compile takeaways from these roundtable discussions, including recent advances, applications, and open challenges for each topic. We conclude with a summary and lessons learned across all roundtables. This document serves as a comprehensive review paper, summarizing the recent advancements in machine learning for healthcare as contributed by foremost researchers in the field.
Zero-Shot Clinical Trial Patient Matching with LLMs
Feb 05, 2024
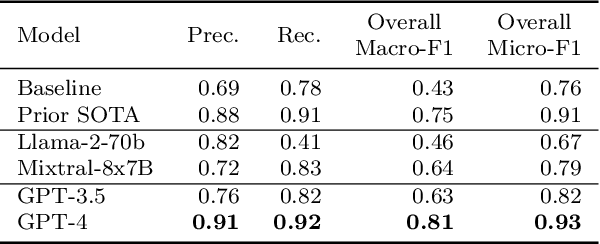
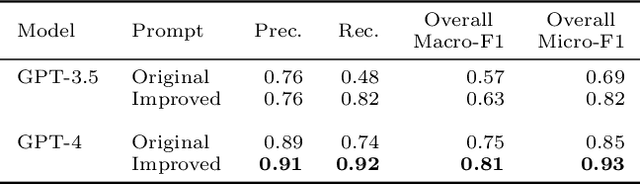

Abstract:Matching patients to clinical trials is a key unsolved challenge in bringing new drugs to market. Today, identifying patients who meet a trial's eligibility criteria is highly manual, taking up to 1 hour per patient. Automated screening is challenging, however, as it requires understanding unstructured clinical text. Large language models (LLMs) offer a promising solution. In this work, we explore their application to trial matching. First, we design an LLM-based system which, given a patient's medical history as unstructured clinical text, evaluates whether that patient meets a set of inclusion criteria (also specified as free text). Our zero-shot system achieves state-of-the-art scores on the n2c2 2018 cohort selection benchmark. Second, we improve the data and cost efficiency of our method by identifying a prompting strategy which matches patients an order of magnitude faster and more cheaply than the status quo, and develop a two-stage retrieval pipeline that reduces the number of tokens processed by up to a third while retaining high performance. Third, we evaluate the interpretability of our system by having clinicians evaluate the natural language justifications generated by the LLM for each eligibility decision, and show that it can output coherent explanations for 97% of its correct decisions and 75% of its incorrect ones. Our results establish the feasibility of using LLMs to accelerate clinical trial operations.
EHRSHOT: An EHR Benchmark for Few-Shot Evaluation of Foundation Models
Jul 05, 2023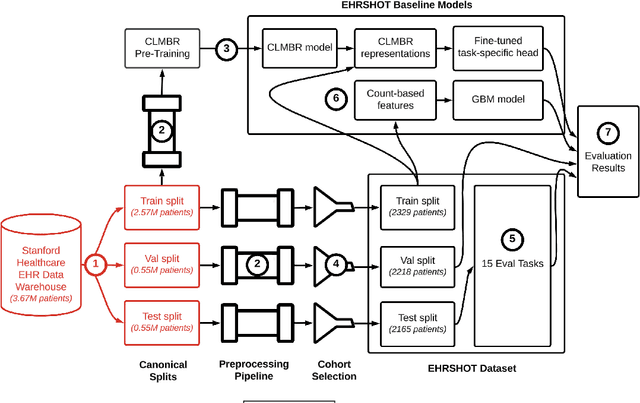
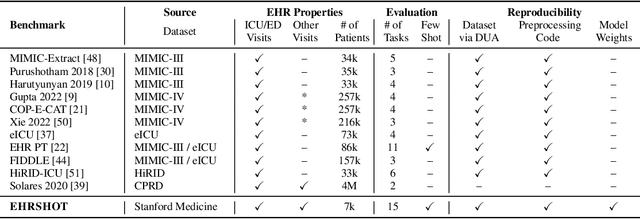

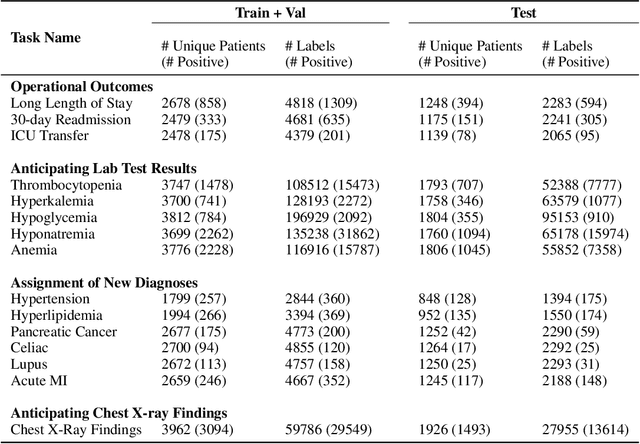
Abstract:While the general machine learning (ML) community has benefited from public datasets, tasks, and models, the progress of ML in healthcare has been hampered by a lack of such shared assets. The success of foundation models creates new challenges for healthcare ML by requiring access to shared pretrained models to validate performance benefits. We help address these challenges through three contributions. First, we publish a new dataset, EHRSHOT, containing de-identified structured data from the electronic health records (EHRs) of 6,712 patients from Stanford Medicine. Unlike MIMIC-III/IV and other popular EHR datasets, EHRSHOT is longitudinal and not restricted to ICU/ED patients. Second, we publish the weights of a 141M parameter clinical foundation model pretrained on the structured EHR data of 2.57M patients. We are one of the first to fully release such a model for coded EHR data; in contrast, most prior models released for clinical data (e.g. GatorTron, ClinicalBERT) only work with unstructured text and cannot process the rich, structured data within an EHR. We provide an end-to-end pipeline for the community to validate and build upon its performance. Third, we define 15 few-shot clinical prediction tasks, enabling evaluation of foundation models on benefits such as sample efficiency and task adaption. The code to reproduce our results, as well as the model and dataset (via a research data use agreement), are available at our Github repo here: https://github.com/som-shahlab/ehrshot-benchmark
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge