Juan M. Banda
Monitoring Deployed AI Systems in Health Care
Dec 09, 2025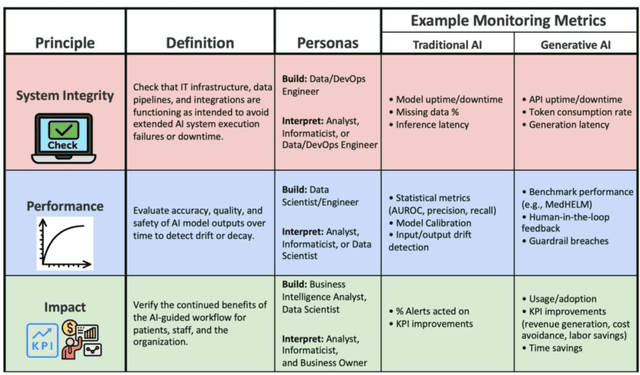
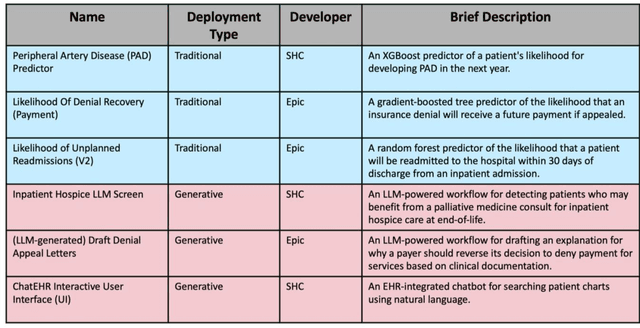
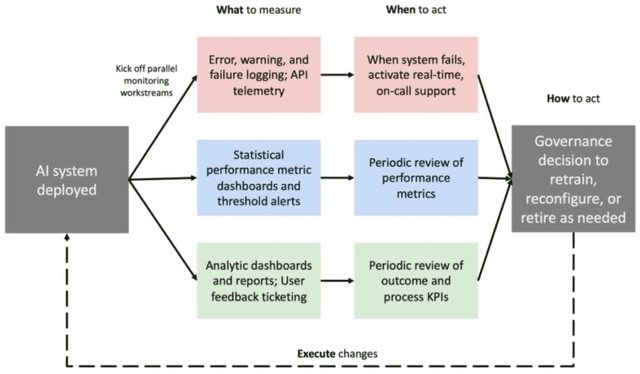
Abstract:Post-deployment monitoring of artificial intelligence (AI) systems in health care is essential to ensure their safety, quality, and sustained benefit-and to support governance decisions about which systems to update, modify, or decommission. Motivated by these needs, we developed a framework for monitoring deployed AI systems grounded in the mandate to take specific actions when they fail to behave as intended. This framework, which is now actively used at Stanford Health Care, is organized around three complementary principles: system integrity, performance, and impact. System integrity monitoring focuses on maximizing system uptime, detecting runtime errors, and identifying when changes to the surrounding IT ecosystem have unintended effects. Performance monitoring focuses on maintaining accurate system behavior in the face of changing health care practices (and thus input data) over time. Impact monitoring assesses whether a deployed system continues to have value in the form of benefit to clinicians and patients. Drawing on examples of deployed AI systems at our academic medical center, we provide practical guidance for creating monitoring plans based on these principles that specify which metrics to measure, when those metrics should be reviewed, who is responsible for acting when metrics change, and what concrete follow-up actions should be taken-for both traditional and generative AI. We also discuss challenges to implementing this framework, including the effort and cost of monitoring for health systems with limited resources and the difficulty of incorporating data-driven monitoring practices into complex organizations where conflicting priorities and definitions of success often coexist. This framework offers a practical template and starting point for health systems seeking to ensure that AI deployments remain safe and effective over time.
MedHELM: Holistic Evaluation of Large Language Models for Medical Tasks
May 26, 2025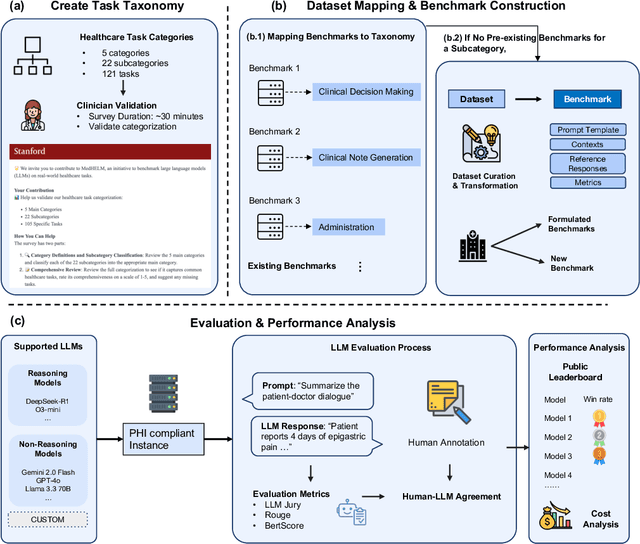
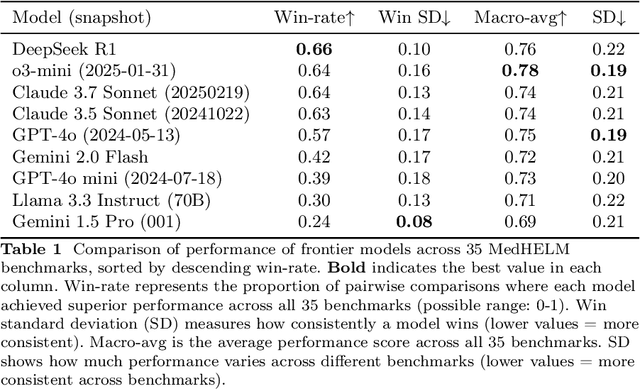
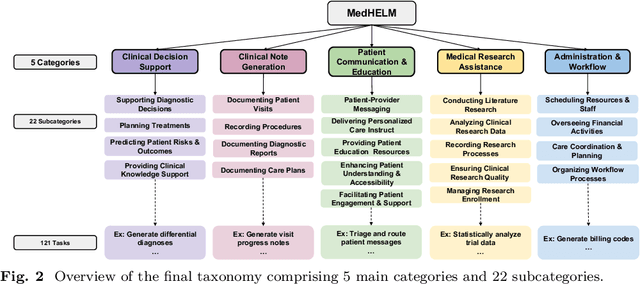
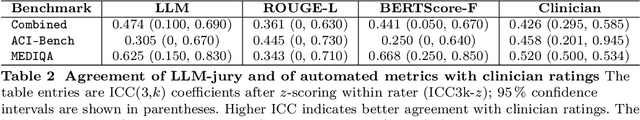
Abstract:While large language models (LLMs) achieve near-perfect scores on medical licensing exams, these evaluations inadequately reflect the complexity and diversity of real-world clinical practice. We introduce MedHELM, an extensible evaluation framework for assessing LLM performance for medical tasks with three key contributions. First, a clinician-validated taxonomy spanning 5 categories, 22 subcategories, and 121 tasks developed with 29 clinicians. Second, a comprehensive benchmark suite comprising 35 benchmarks (17 existing, 18 newly formulated) providing complete coverage of all categories and subcategories in the taxonomy. Third, a systematic comparison of LLMs with improved evaluation methods (using an LLM-jury) and a cost-performance analysis. Evaluation of 9 frontier LLMs, using the 35 benchmarks, revealed significant performance variation. Advanced reasoning models (DeepSeek R1: 66% win-rate; o3-mini: 64% win-rate) demonstrated superior performance, though Claude 3.5 Sonnet achieved comparable results at 40% lower estimated computational cost. On a normalized accuracy scale (0-1), most models performed strongly in Clinical Note Generation (0.73-0.85) and Patient Communication & Education (0.78-0.83), moderately in Medical Research Assistance (0.65-0.75), and generally lower in Clinical Decision Support (0.56-0.72) and Administration & Workflow (0.53-0.63). Our LLM-jury evaluation method achieved good agreement with clinician ratings (ICC = 0.47), surpassing both average clinician-clinician agreement (ICC = 0.43) and automated baselines including ROUGE-L (0.36) and BERTScore-F1 (0.44). Claude 3.5 Sonnet achieved comparable performance to top models at lower estimated cost. These findings highlight the importance of real-world, task-specific evaluation for medical use of LLMs and provides an open source framework to enable this.
Standing on FURM ground -- A framework for evaluating Fair, Useful, and Reliable AI Models in healthcare systems
Mar 14, 2024



Abstract:The impact of using artificial intelligence (AI) to guide patient care or operational processes is an interplay of the AI model's output, the decision-making protocol based on that output, and the capacity of the stakeholders involved to take the necessary subsequent action. Estimating the effects of this interplay before deployment, and studying it in real time afterwards, are essential to bridge the chasm between AI model development and achievable benefit. To accomplish this, the Data Science team at Stanford Health Care has developed a Testing and Evaluation (T&E) mechanism to identify fair, useful and reliable AI models (FURM) by conducting an ethical review to identify potential value mismatches, simulations to estimate usefulness, financial projections to assess sustainability, as well as analyses to determine IT feasibility, design a deployment strategy, and recommend a prospective monitoring and evaluation plan. We report on FURM assessments done to evaluate six AI guided solutions for potential adoption, spanning clinical and operational settings, each with the potential to impact from several dozen to tens of thousands of patients each year. We describe the assessment process, summarize the six assessments, and share our framework to enable others to conduct similar assessments. Of the six solutions we assessed, two have moved into a planning and implementation phase. Our novel contributions - usefulness estimates by simulation, financial projections to quantify sustainability, and a process to do ethical assessments - as well as their underlying methods and open source tools, are available for other healthcare systems to conduct actionable evaluations of candidate AI solutions.
Leveraging Large Language Models and Weak Supervision for Social Media data annotation: an evaluation using COVID-19 self-reported vaccination tweets
Sep 12, 2023
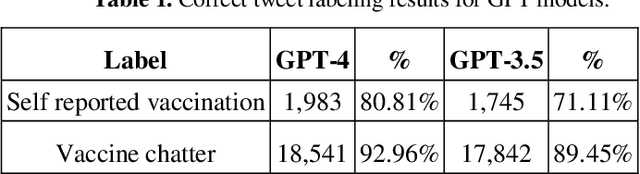

Abstract:The COVID-19 pandemic has presented significant challenges to the healthcare industry and society as a whole. With the rapid development of COVID-19 vaccines, social media platforms have become a popular medium for discussions on vaccine-related topics. Identifying vaccine-related tweets and analyzing them can provide valuable insights for public health research-ers and policymakers. However, manual annotation of a large number of tweets is time-consuming and expensive. In this study, we evaluate the usage of Large Language Models, in this case GPT-4 (March 23 version), and weak supervision, to identify COVID-19 vaccine-related tweets, with the purpose of comparing performance against human annotators. We leveraged a manu-ally curated gold-standard dataset and used GPT-4 to provide labels without any additional fine-tuning or instructing, in a single-shot mode (no additional prompting).
Evaluation of GPT-3.5 and GPT-4 for supporting real-world information needs in healthcare delivery
May 01, 2023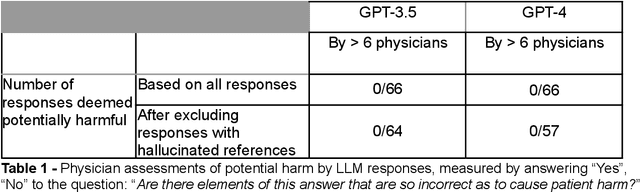
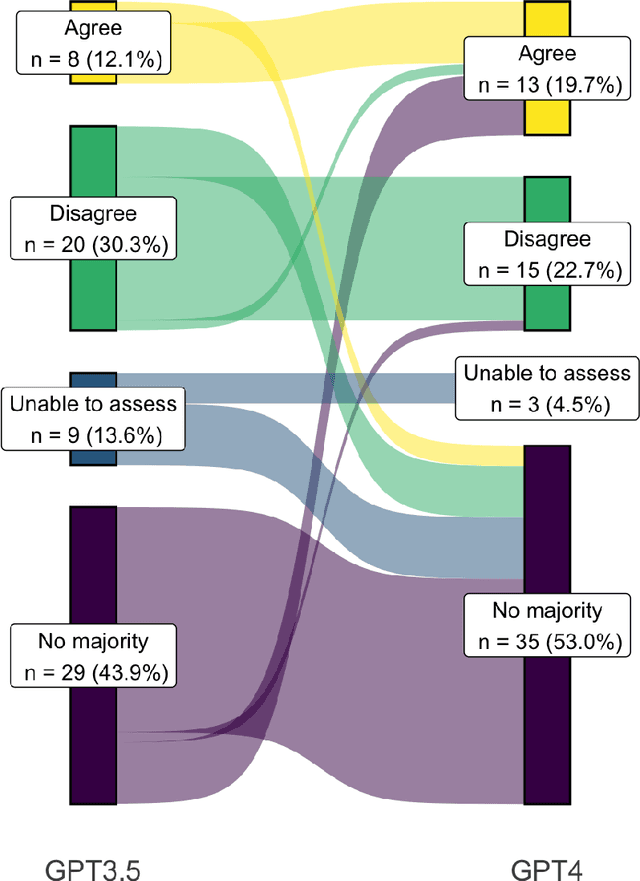
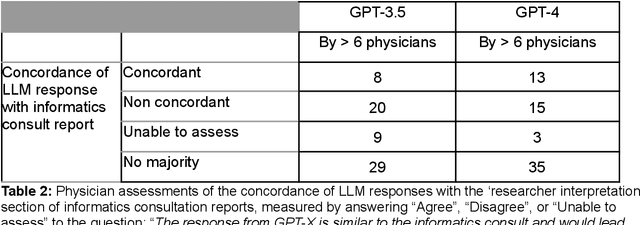
Abstract:Despite growing interest in using large language models (LLMs) in healthcare, current explorations do not assess the real-world utility and safety of LLMs in clinical settings. Our objective was to determine whether two LLMs can serve information needs submitted by physicians as questions to an informatics consultation service in a safe and concordant manner. Sixty six questions from an informatics consult service were submitted to GPT-3.5 and GPT-4 via simple prompts. 12 physicians assessed the LLM responses' possibility of patient harm and concordance with existing reports from an informatics consultation service. Physician assessments were summarized based on majority vote. For no questions did a majority of physicians deem either LLM response as harmful. For GPT-3.5, responses to 8 questions were concordant with the informatics consult report, 20 discordant, and 9 were unable to be assessed. There were 29 responses with no majority on "Agree", "Disagree", and "Unable to assess". For GPT-4, responses to 13 questions were concordant, 15 discordant, and 3 were unable to be assessed. There were 35 responses with no majority. Responses from both LLMs were largely devoid of overt harm, but less than 20% of the responses agreed with an answer from an informatics consultation service, responses contained hallucinated references, and physicians were divided on what constitutes harm. These results suggest that while general purpose LLMs are able to provide safe and credible responses, they often do not meet the specific information need of a given question. A definitive evaluation of the usefulness of LLMs in healthcare settings will likely require additional research on prompt engineering, calibration, and custom-tailoring of general purpose models.
Ontologizing Health Systems Data at Scale: Making Translational Discovery a Reality
Sep 10, 2022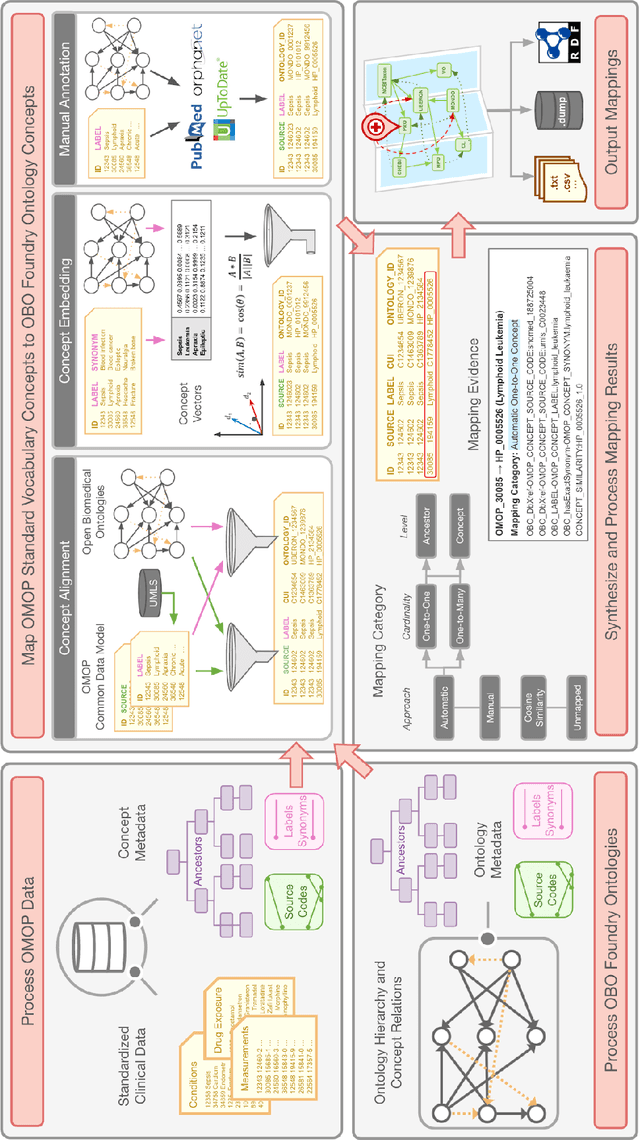
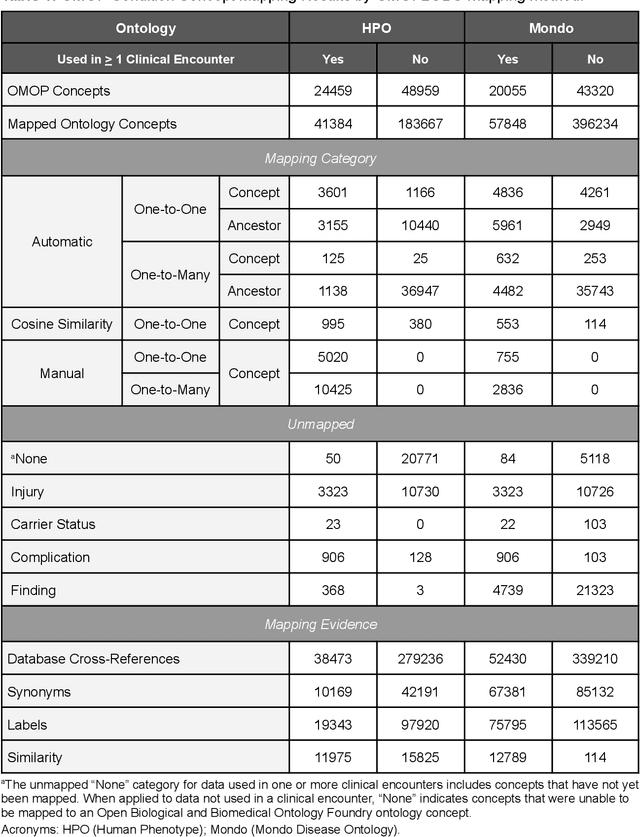
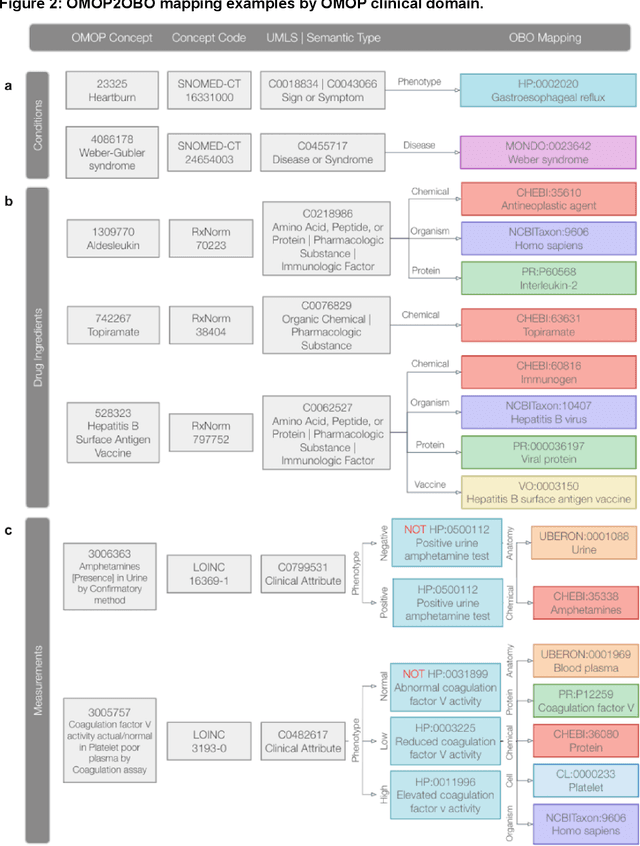
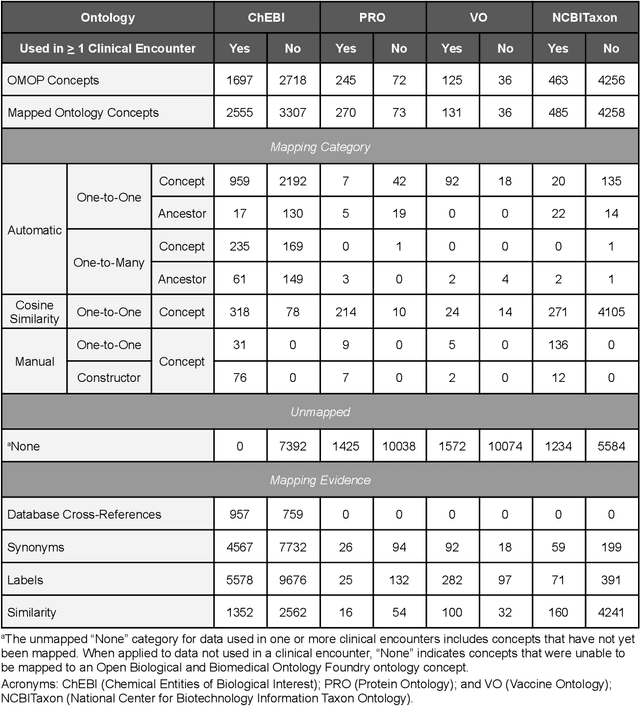
Abstract:Common data models solve many challenges of standardizing electronic health record (EHR) data, but are unable to semantically integrate the resources needed for deep phenotyping. Open Biological and Biomedical Ontology (OBO) Foundry ontologies provide semantically computable representations of biological knowledge and enable the integration of a variety of biomedical data. However, mapping EHR data to OBO Foundry ontologies requires significant manual curation and domain expertise. We introduce a framework for mapping Observational Medical Outcomes Partnership (OMOP) standard vocabularies to OBO Foundry ontologies. Using this framework, we produced mappings for 92,367 conditions, 8,615 drug ingredients, and 10,673 measurement results. Mapping accuracy was verified by domain experts and when examined across 24 hospitals, the mappings covered 99% of conditions and drug ingredients and 68% of measurements. Finally, we demonstrate that OMOP2OBO mappings can aid in the systematic identification of undiagnosed rare disease patients who might benefit from genetic testing.
TweetDIS: A Large Twitter Dataset for Natural Disasters Built using Weak Supervision
Jul 11, 2022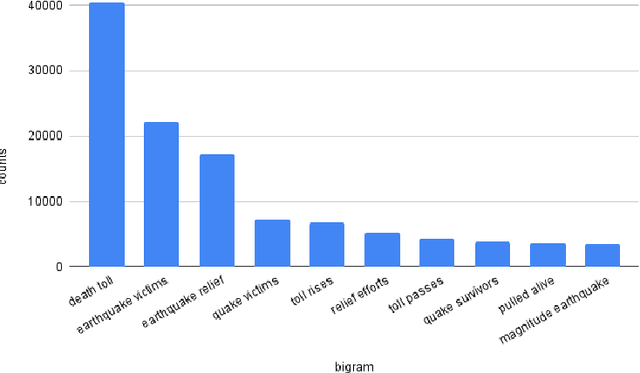
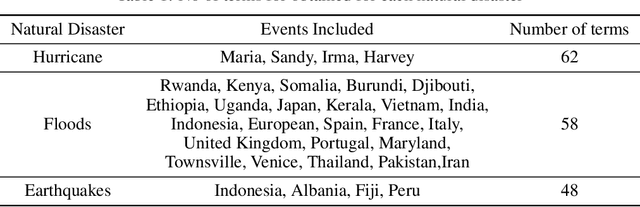
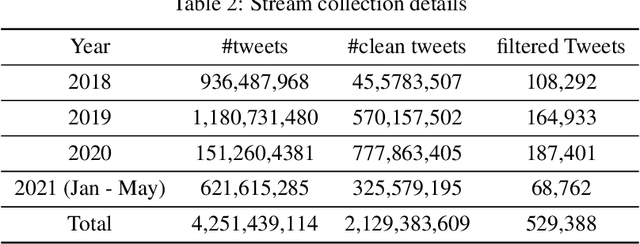
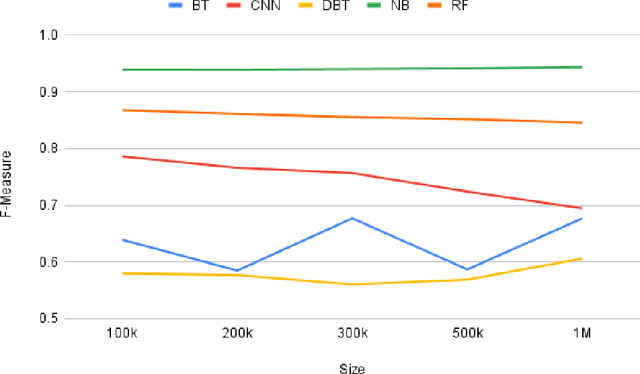
Abstract:Social media is often utilized as a lifeline for communication during natural disasters. Traditionally, natural disaster tweets are filtered from the Twitter stream using the name of the natural disaster and the filtered tweets are sent for human annotation. The process of human annotation to create labeled sets for machine learning models is laborious, time consuming, at times inaccurate, and more importantly not scalable in terms of size and real-time use. In this work, we curate a silver standard dataset using weak supervision. In order to validate its utility, we train machine learning models on the weakly supervised data to identify three different types of natural disasters i.e earthquakes, hurricanes and floods. Our results demonstrate that models trained on the silver standard dataset achieved performance greater than 90% when classifying a manually curated, gold-standard dataset. To enable reproducible research and additional downstream utility, we release the silver standard dataset for the scientific community.
A Biomedically oriented automatically annotated Twitter COVID-19 Dataset
Jul 27, 2021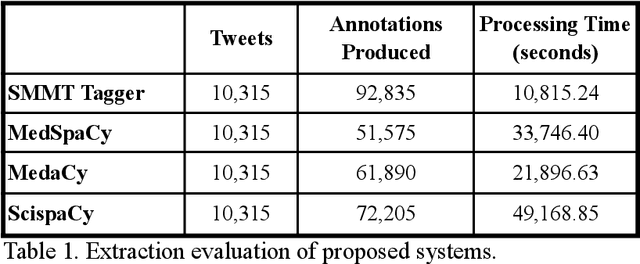
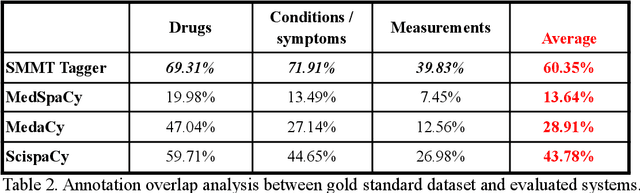
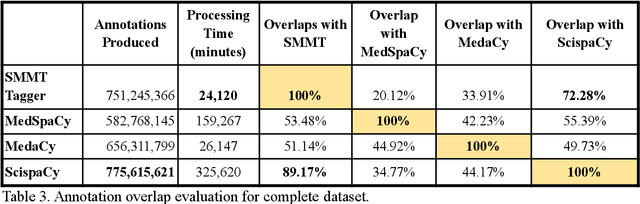
Abstract:The use of social media data, like Twitter, for biomedical research has been gradually increasing over the years. With the COVID-19 pandemic, researchers have turned to more nontraditional sources of clinical data to characterize the disease in near real-time, study the societal implications of interventions, as well as the sequelae that recovered COVID-19 cases present (Long-COVID). However, manually curated social media datasets are difficult to come by due to the expensive costs of manual annotation and the efforts needed to identify the correct texts. When datasets are available, they are usually very small and their annotations do not generalize well over time or to larger sets of documents. As part of the 2021 Biomedical Linked Annotation Hackathon, we release our dataset of over 120 million automatically annotated tweets for biomedical research purposes. Incorporating best practices, we identify tweets with potentially high clinical relevance. We evaluated our work by comparing several SpaCy-based annotation frameworks against a manually annotated gold-standard dataset. Selecting the best method to use for automatic annotation, we then annotated 120 million tweets and released them publicly for future downstream usage within the biomedical domain.
Characterization of Potential Drug Treatments for COVID-19 using Social Media Data and Machine Learning
Jul 20, 2020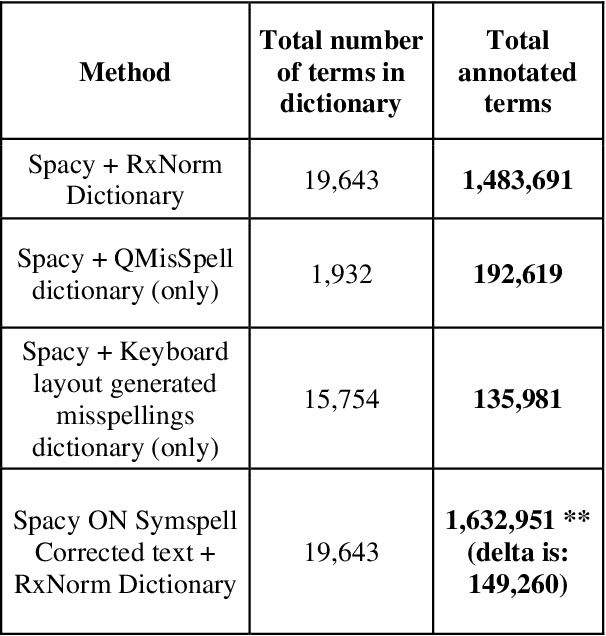
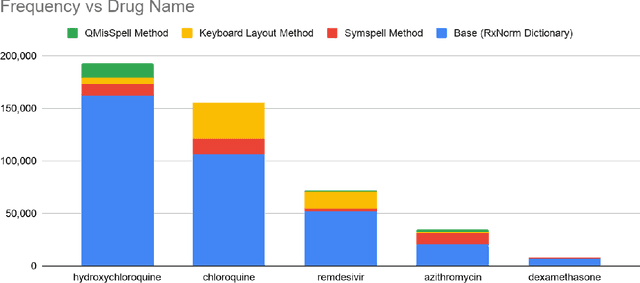
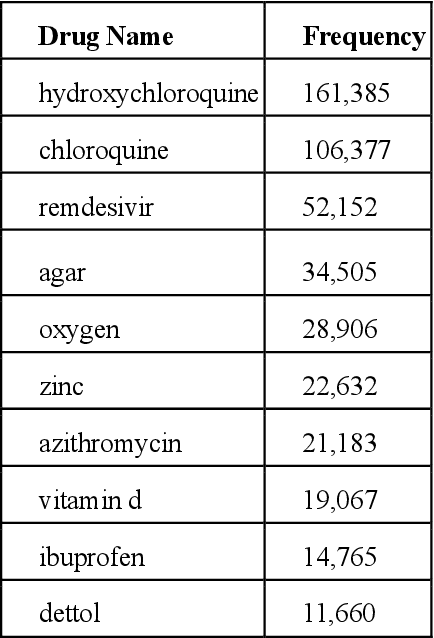
Abstract:Since the classification of COVID-19 as a global pandemic, there have been many attempts to treat and contain the virus. Although there is no specific antiviral treatment recommended for COVID-19, there are several drugs that can potentially help with symptoms. In this work, we mined a large twitter dataset of 424 million tweets of COVID-19 chatter to identify discourse around potential treatments. While seemingly a straightforward task, due to the informal nature of language use in Twitter, we demonstrate the need of machine learning methods to aid in this task. By applying these methods we are able to recover almost 15% additional data than with traditional methods, showing the need of more sophisticated approaches than just text matching.
GLEAKE: Global and Local Embedding Automatic Keyphrase Extraction
May 19, 2020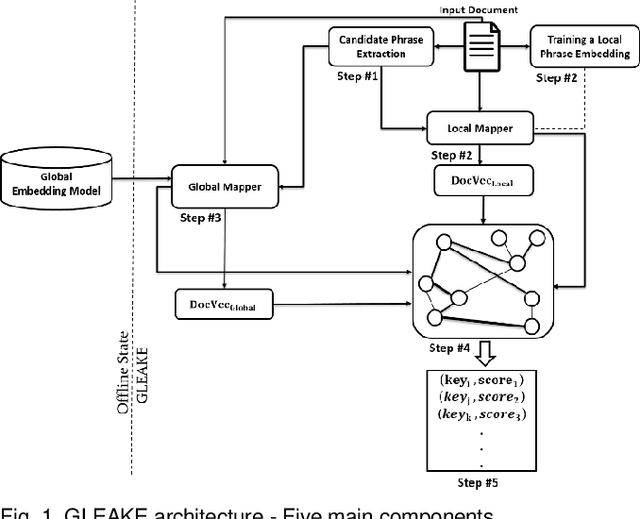
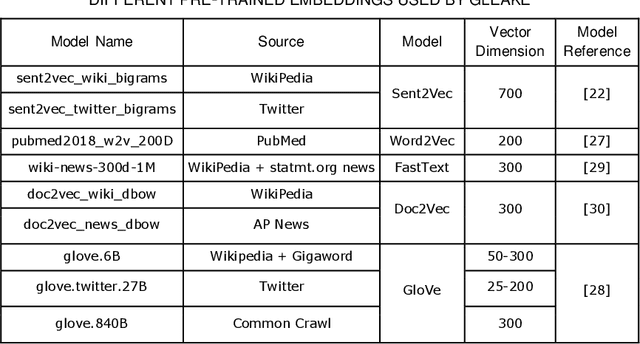
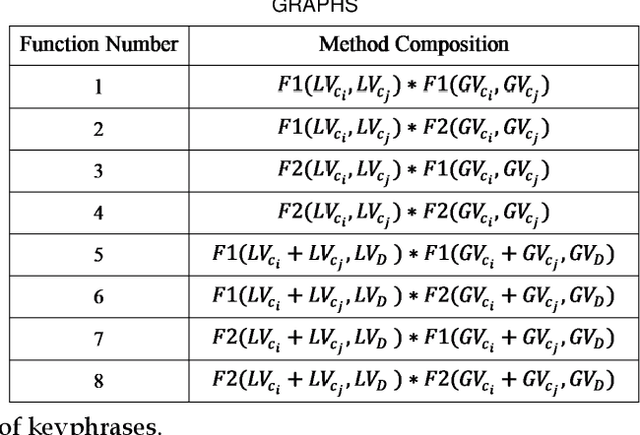
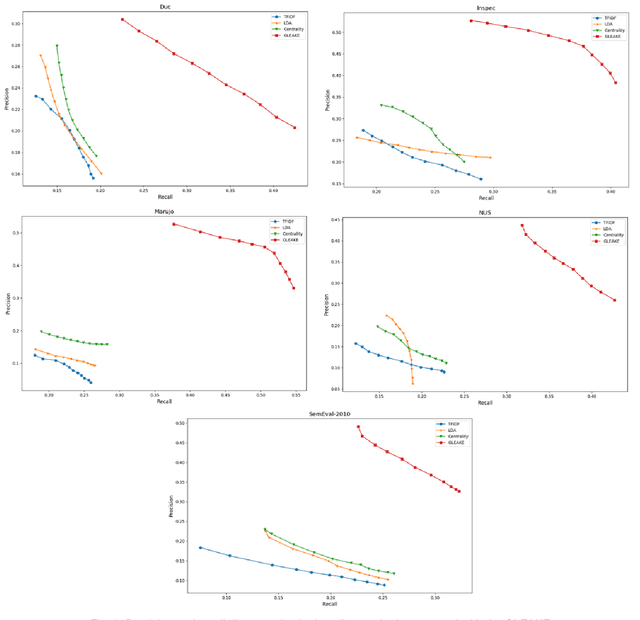
Abstract:Automated methods for granular categorization of large corpora of text documents have become increasingly more important with the rate scientific, news, medical, and web documents are growing in the last few years. Automatic keyphrase extraction (AKE) aims to automatically detect a small set of single or multi-words from within a single textual document that captures the main topics of the document. AKE plays an important role in various NLP and information retrieval tasks such as document summarization and categorization, full-text indexing, and article recommendation. Due to the lack of sufficient human-labeled data in different textual contents, supervised learning approaches are not ideal for automatic detection of keyphrases from the content of textual bodies. With the state-of-the-art advances in text embedding techniques, NLP researchers have focused on developing unsupervised methods to obtain meaningful insights from raw datasets. In this work, we introduce Global and Local Embedding Automatic Keyphrase Extractor (GLEAKE) for the task of AKE. GLEAKE utilizes single and multi-word embedding techniques to explore the syntactic and semantic aspects of the candidate phrases and then combines them into a series of embedding-based graphs. Moreover, GLEAKE applies network analysis techniques on each embedding-based graph to refine the most significant phrases as a final set of keyphrases. We demonstrate the high performance of GLEAKE by evaluating its results on five standard AKE datasets from different domains and writing styles and by showing its superiority with regards to other state-of-the-art methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge