Elena Casiraghi
on behalf of the N3C Consortium
Interpreting Manifolds and Graph Neural Embeddings from Internet of Things Traffic Flows
Feb 05, 2026Abstract:The rapid expansion of Internet of Things (IoT) ecosystems has led to increasingly complex and heterogeneous network topologies. Traditional network monitoring and visualization tools rely on aggregated metrics or static representations, which fail to capture the evolving relationships and structural dependencies between devices. Although Graph Neural Networks (GNNs) offer a powerful way to learn from relational data, their internal representations often remain opaque and difficult to interpret for security-critical operations. Consequently, this work introduces an interpretable pipeline that generates directly visualizable low-dimensional representations by mapping high-dimensional embeddings onto a latent manifold. This projection enables the interpretable monitoring and interoperability of evolving network states, while integrated feature attribution techniques decode the specific characteristics shaping the manifold structure. The framework achieves a classification F1-score of 0.830 for intrusion detection while also highlighting phenomena such as concept drift. Ultimately, the presented approach bridges the gap between high-dimensional GNN embeddings and human-understandable network behavior, offering new insights for network administrators and security analysts.
RNA-KG: An ontology-based knowledge graph for representing interactions involving RNA molecules
Nov 30, 2023Abstract:The "RNA world" represents a novel frontier for the study of fundamental biological processes and human diseases and is paving the way for the development of new drugs tailored to the patient's biomolecular characteristics. Although scientific data about coding and non-coding RNA molecules are continuously produced and available from public repositories, they are scattered across different databases and a centralized, uniform, and semantically consistent representation of the "RNA world" is still lacking. We propose RNA-KG, a knowledge graph encompassing biological knowledge about RNAs gathered from more than 50 public databases, integrating functional relationships with genes, proteins, and chemicals and ontologically grounded biomedical concepts. To develop RNA-KG, we first identified, pre-processed, and characterized each data source; next, we built a meta-graph that provides an ontological description of the KG by representing all the bio-molecular entities and medical concepts of interest in this domain, as well as the types of interactions connecting them. Finally, we leveraged an instance-based semantically abstracted knowledge model to specify the ontological alignment according to which RNA-KG was generated. RNA-KG can be downloaded in different formats and also queried by a SPARQL endpoint. A thorough topological analysis of the resulting heterogeneous graph provides further insights into the characteristics of the "RNA world". RNA-KG can be both directly explored and visualized, and/or analyzed by applying computational methods to infer bio-medical knowledge from its heterogeneous nodes and edges. The resource can be easily updated with new experimental data, and specific views of the overall KG can be extracted according to the bio-medical problem to be studied.
An Open-Source Knowledge Graph Ecosystem for the Life Sciences
Jul 11, 2023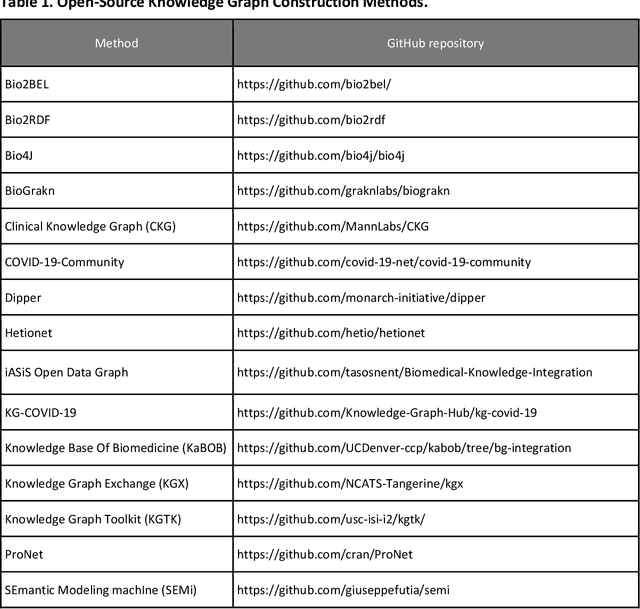
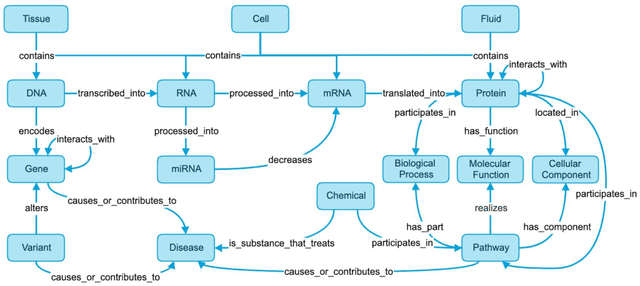
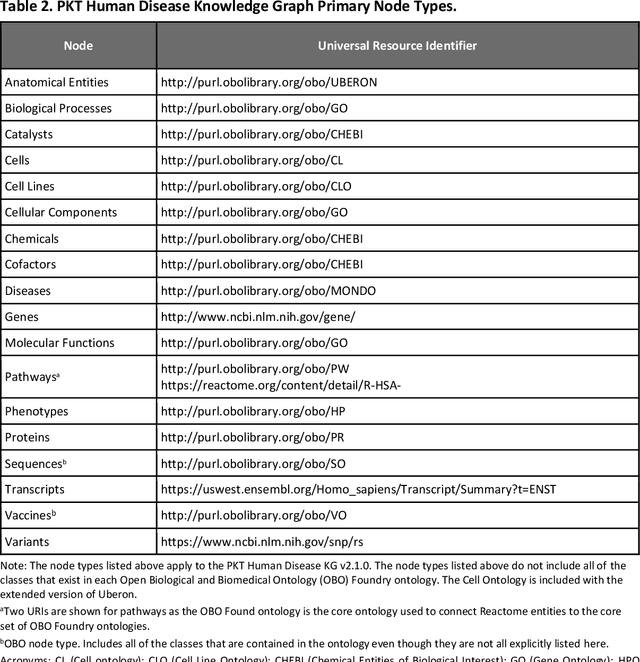
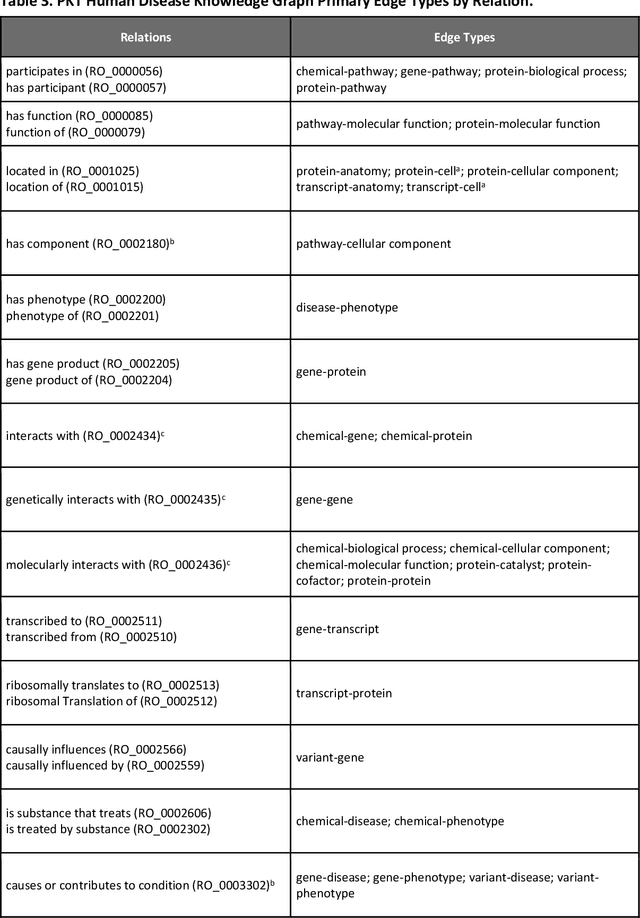
Abstract:Translational research requires data at multiple scales of biological organization. Advancements in sequencing and multi-omics technologies have increased the availability of these data but researchers face significant integration challenges. Knowledge graphs (KGs) are used to model complex phenomena, and methods exist to automatically construct them. However, tackling complex biomedical integration problems requires flexibility in the way knowledge is modeled. Moreover, existing KG construction methods provide robust tooling at the cost of fixed or limited choices among knowledge representation models. PheKnowLator (Phenotype Knowledge Translator) is a semantic ecosystem for automating the FAIR (Findable, Accessible, Interoperable, and Reusable) construction of ontologically grounded KGs with fully customizable knowledge representation. The ecosystem includes KG construction resources (e.g., data preparation APIs), analysis tools (e.g., SPARQL endpoints and abstraction algorithms), and benchmarks (e.g., prebuilt KGs and embeddings). We evaluate the ecosystem by surveying open-source KG construction methods and analyzing its computational performance when constructing 12 large-scale KGs. With flexible knowledge representation, PheKnowLator enables fully customizable KGs without compromising performance or usability.
Ontologizing Health Systems Data at Scale: Making Translational Discovery a Reality
Sep 10, 2022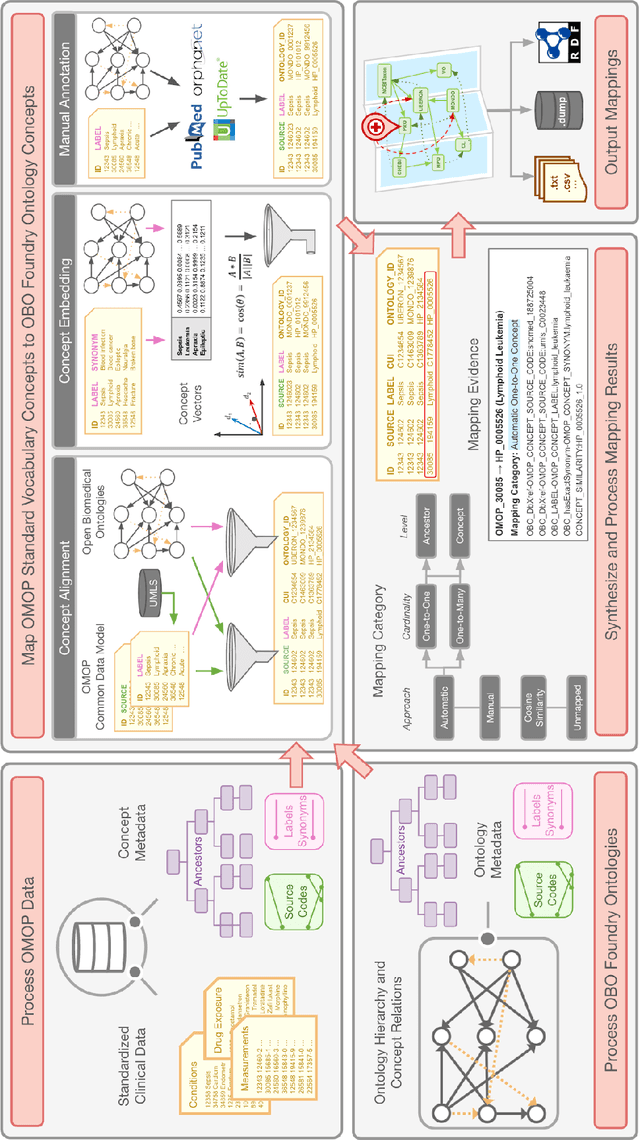
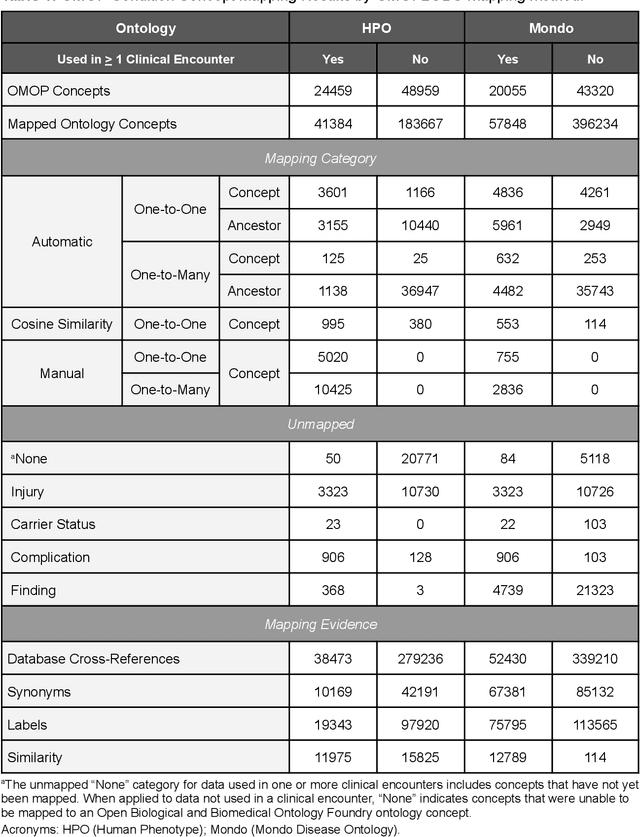
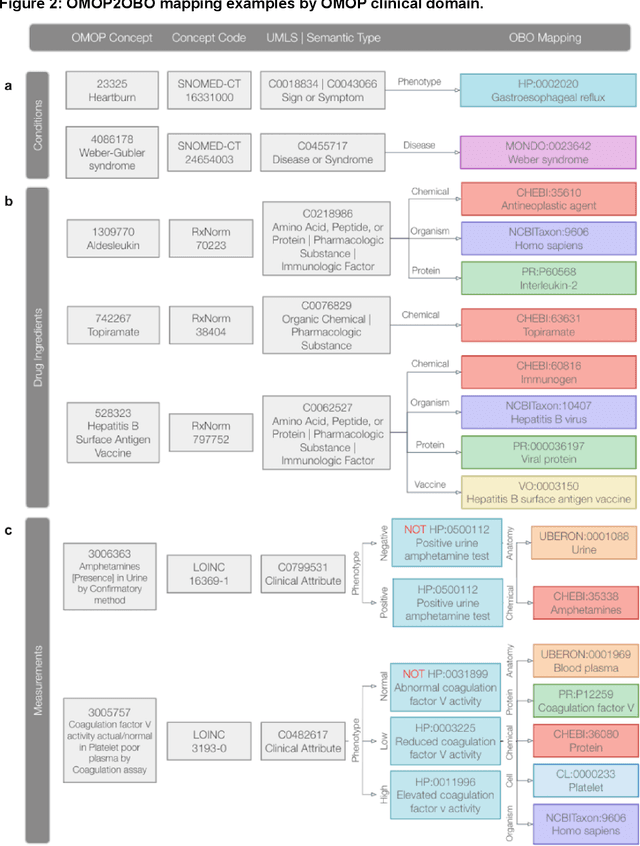
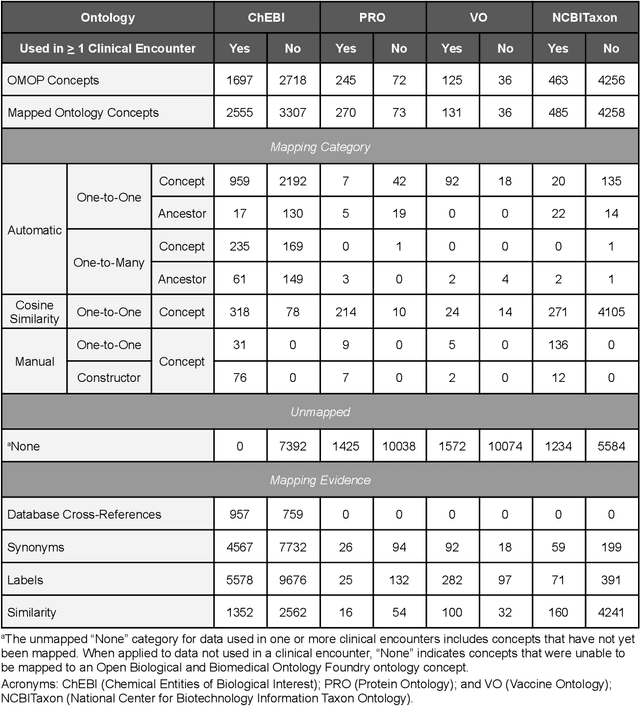
Abstract:Common data models solve many challenges of standardizing electronic health record (EHR) data, but are unable to semantically integrate the resources needed for deep phenotyping. Open Biological and Biomedical Ontology (OBO) Foundry ontologies provide semantically computable representations of biological knowledge and enable the integration of a variety of biomedical data. However, mapping EHR data to OBO Foundry ontologies requires significant manual curation and domain expertise. We introduce a framework for mapping Observational Medical Outcomes Partnership (OMOP) standard vocabularies to OBO Foundry ontologies. Using this framework, we produced mappings for 92,367 conditions, 8,615 drug ingredients, and 10,673 measurement results. Mapping accuracy was verified by domain experts and when examined across 24 hospitals, the mappings covered 99% of conditions and drug ingredients and 68% of measurements. Finally, we demonstrate that OMOP2OBO mappings can aid in the systematic identification of undiagnosed rare disease patients who might benefit from genetic testing.
A Methodological Framework for the Comparative Evaluation of Multiple Imputation Methods: Multiple Imputation of Race, Ethnicity and Body Mass Index in the U.S. National COVID Cohort Collaborative
Jun 13, 2022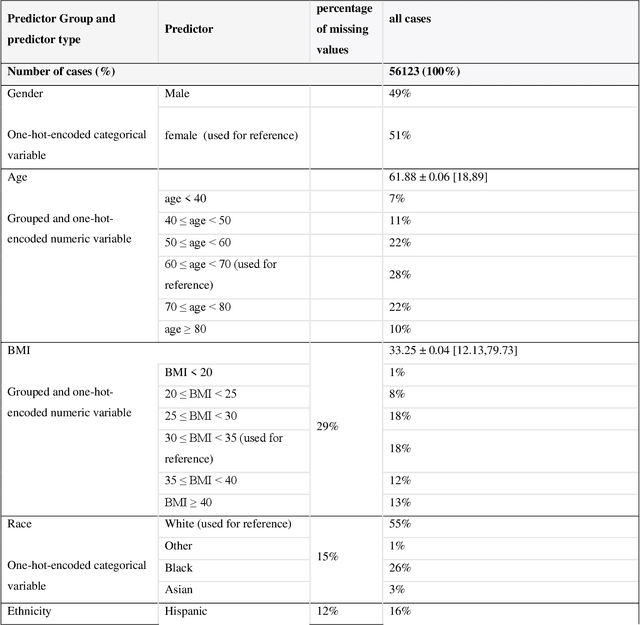
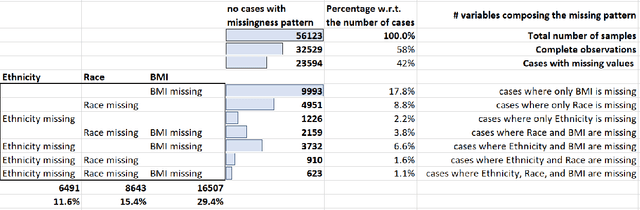
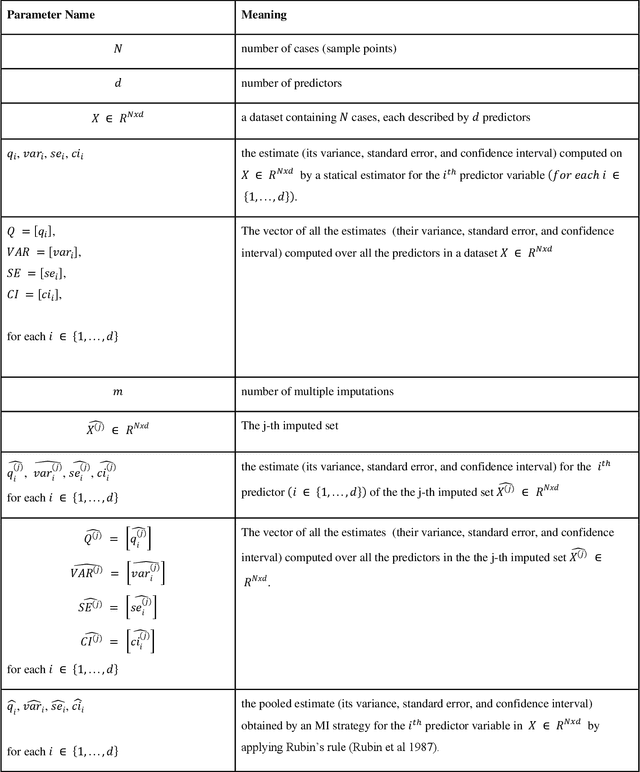
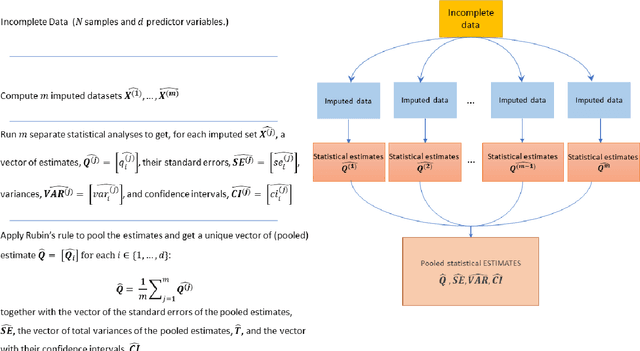
Abstract:While electronic health records are a rich data source for biomedical research, these systems are not implemented uniformly across healthcare settings and significant data may be missing due to healthcare fragmentation and lack of interoperability between siloed electronic health records. Considering that the deletion of cases with missing data may introduce severe bias in the subsequent analysis, several authors prefer applying a multiple imputation strategy to recover the missing information. Unfortunately, although several literature works have documented promising results by using any of the different multiple imputation algorithms that are now freely available for research, there is no consensus on which MI algorithm works best. Beside the choice of the MI strategy, the choice of the imputation algorithm and its application settings are also both crucial and challenging. In this paper, inspired by the seminal works of Rubin and van Buuren, we propose a methodological framework that may be applied to evaluate and compare several multiple imputation techniques, with the aim to choose the most valid for computing inferences in a clinical research work. Our framework has been applied to validate, and extend on a larger cohort, the results we presented in a previous literature study, where we evaluated the influence of crucial patients' descriptors and COVID-19 severity in patients with type 2 diabetes mellitus whose data is provided by the National COVID Cohort Collaborative Enclave.
GraPE: fast and scalable Graph Processing and Embedding
Oct 12, 2021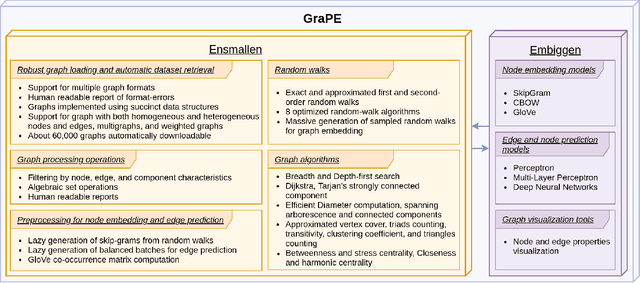
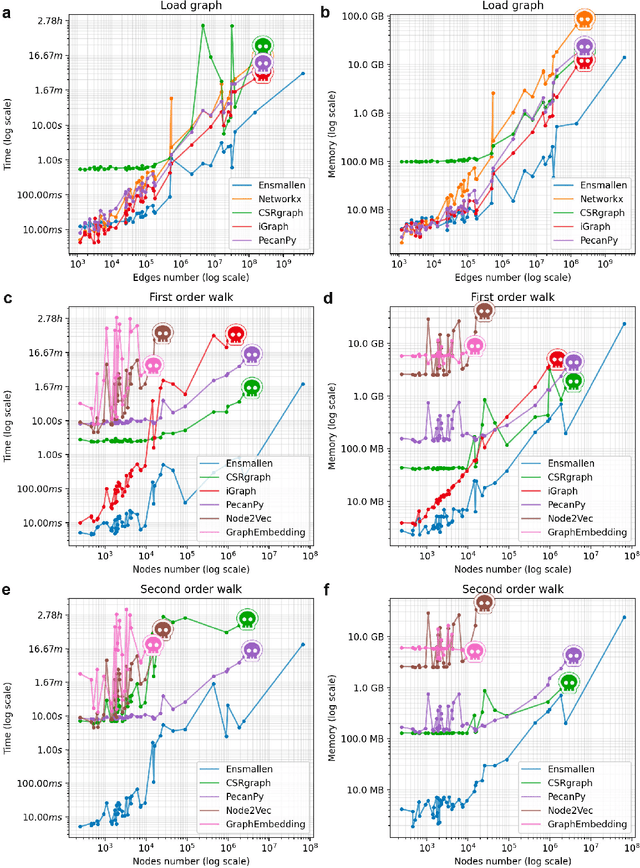
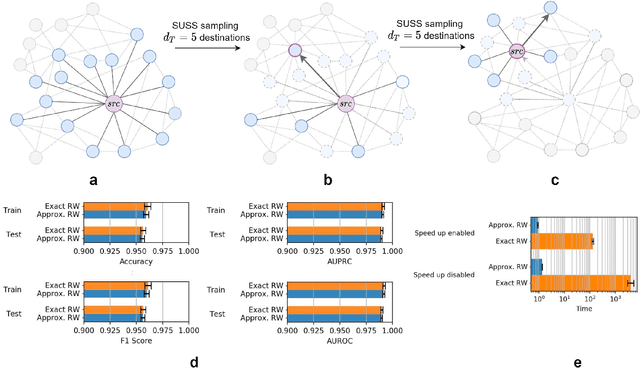
Abstract:Graph Representation Learning methods have enabled a wide range of learning problems to be addressed for data that can be represented in graph form. Nevertheless, several real world problems in economy, biology, medicine and other fields raised relevant scaling problems with existing methods and their software implementation, due to the size of real world graphs characterized by millions of nodes and billions of edges. We present GraPE, a software resource for graph processing and random walk based embedding, that can scale with large and high-degree graphs and significantly speed up-computation. GraPE comprises specialized data structures, algorithms, and a fast parallel implementation that displays everal orders of magnitude improvement in empirical space and time complexity compared to state of the art software resources, with a corresponding boost in the performance of machine learning methods for edge and node label prediction and for the unsupervised analysis of graphs.GraPE is designed to run on laptop and desktop computers, as well as on high performance computing clusters
Het-node2vec: second order random walk sampling for heterogeneous multigraphs embedding
Jan 05, 2021



Abstract:We introduce a set of algorithms (Het-node2vec) that extend the original node2vec node-neighborhood sampling method to heterogeneous multigraphs, i.e. networks characterized by multiple types of nodes and edges. The resulting random walk samples capture both the structural characteristics of the graph and the semantics of the different types of nodes and edges. The proposed algorithms can focus their attention on specific node or edge types, allowing accurate representations also for underrepresented types of nodes/edges that are of interest for the prediction problem under investigation. These rich and well-focused representations can boost unsupervised and supervised learning on heterogeneous graphs.
DANCo: Dimensionality from Angle and Norm Concentration
Jun 18, 2012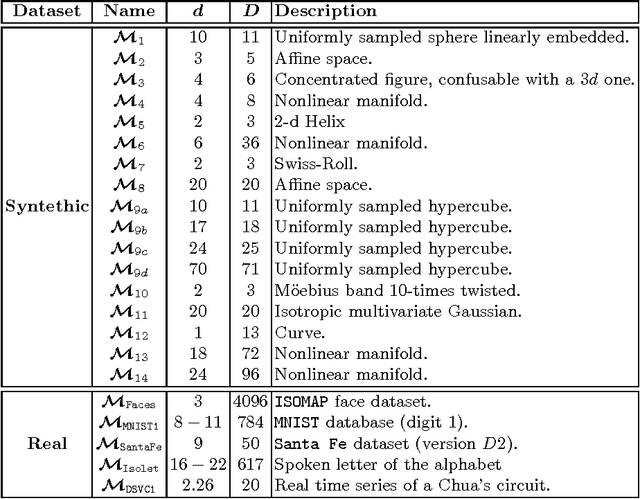
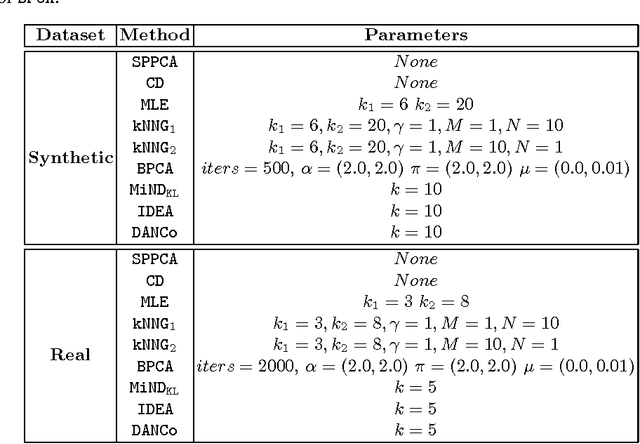
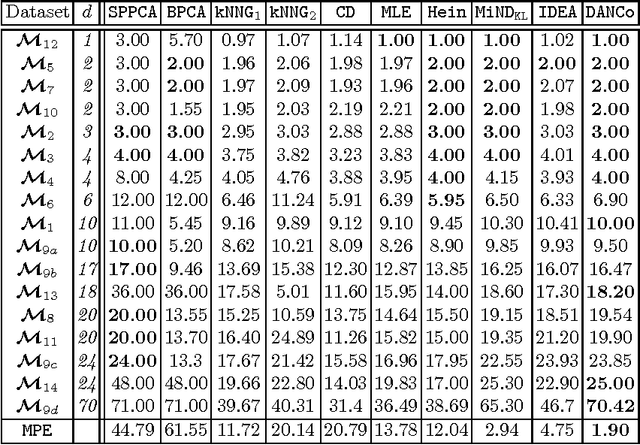
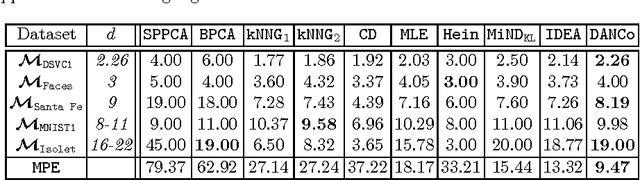
Abstract:In the last decades the estimation of the intrinsic dimensionality of a dataset has gained considerable importance. Despite the great deal of research work devoted to this task, most of the proposed solutions prove to be unreliable when the intrinsic dimensionality of the input dataset is high and the manifold where the points lie is nonlinearly embedded in a higher dimensional space. In this paper we propose a novel robust intrinsic dimensionality estimator that exploits the twofold complementary information conveyed both by the normalized nearest neighbor distances and by the angles computed on couples of neighboring points, providing also closed-forms for the Kullback-Leibler divergences of the respective distributions. Experiments performed on both synthetic and real datasets highlight the robustness and the effectiveness of the proposed algorithm when compared to state of the art methodologies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge