Alberto Cabri
Real-Time Detection of Electronic Components in Waste Printed Circuit Boards: A Transformer-Based Approach
Sep 24, 2024

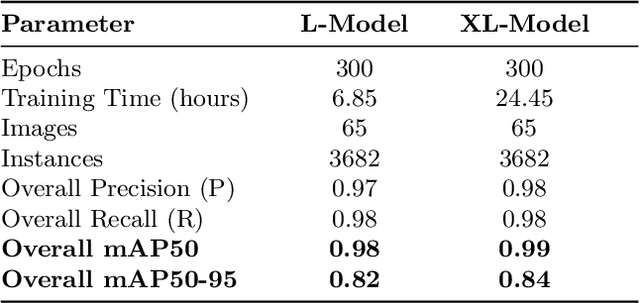
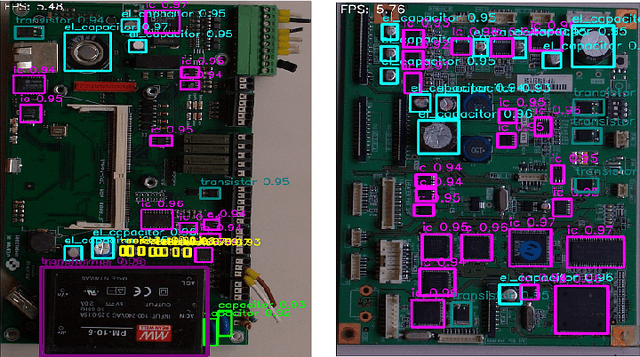
Abstract:Critical Raw Materials (CRMs) such as copper, manganese, gallium, and various rare earths have great importance for the electronic industry. To increase the concentration of individual CRMs and thus make their extraction from Waste Printed Circuit Boards (WPCBs) convenient, we have proposed a practical approach that involves selective disassembling of the different types of electronic components from WPCBs using mechatronic systems guided by artificial vision techniques. In this paper we evaluate the real-time accuracy of electronic component detection and localization of the Real-Time DEtection TRansformer model architecture. Transformers have recently become very popular for the extraordinary results obtained in natural language processing and machine translation. Also in this case, the transformer model achieves very good performances, often superior to those of the latest state of the art object detection and localization models YOLOv8 and YOLOv9.
Virtual Mines -- Component-level recycling of printed circuit boards using deep learning
Jun 24, 2024Abstract:This contribution gives an overview of an ongoing project using machine learning and computer vision components for improving the electronic waste recycling process. In circular economy, the "virtual mines" concept refers to production cycles where interesting raw materials are reclaimed in an efficient and cost-effective manner from end-of-life items. In particular, the growth of e-waste, due to the increasingly shorter life cycle of hi-tech goods, is a global problem. In this paper, we describe a pipeline based on deep learning model to recycle printed circuit boards at the component level. A pre-trained YOLOv5 model is used to analyze the results of the locally developed dataset. With a different distribution of class instances, YOLOv5 managed to achieve satisfactory precision and recall, with the ability to optimize with large component instances.
RNA-KG: An ontology-based knowledge graph for representing interactions involving RNA molecules
Nov 30, 2023Abstract:The "RNA world" represents a novel frontier for the study of fundamental biological processes and human diseases and is paving the way for the development of new drugs tailored to the patient's biomolecular characteristics. Although scientific data about coding and non-coding RNA molecules are continuously produced and available from public repositories, they are scattered across different databases and a centralized, uniform, and semantically consistent representation of the "RNA world" is still lacking. We propose RNA-KG, a knowledge graph encompassing biological knowledge about RNAs gathered from more than 50 public databases, integrating functional relationships with genes, proteins, and chemicals and ontologically grounded biomedical concepts. To develop RNA-KG, we first identified, pre-processed, and characterized each data source; next, we built a meta-graph that provides an ontological description of the KG by representing all the bio-molecular entities and medical concepts of interest in this domain, as well as the types of interactions connecting them. Finally, we leveraged an instance-based semantically abstracted knowledge model to specify the ontological alignment according to which RNA-KG was generated. RNA-KG can be downloaded in different formats and also queried by a SPARQL endpoint. A thorough topological analysis of the resulting heterogeneous graph provides further insights into the characteristics of the "RNA world". RNA-KG can be both directly explored and visualized, and/or analyzed by applying computational methods to infer bio-medical knowledge from its heterogeneous nodes and edges. The resource can be easily updated with new experimental data, and specific views of the overall KG can be extracted according to the bio-medical problem to be studied.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge