Anna Ostropolets
What are the Desired Characteristics of Calibration Sets? Identifying Correlates on Long Form Scientific Summarization
May 12, 2023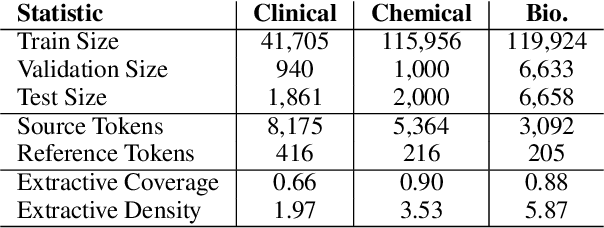
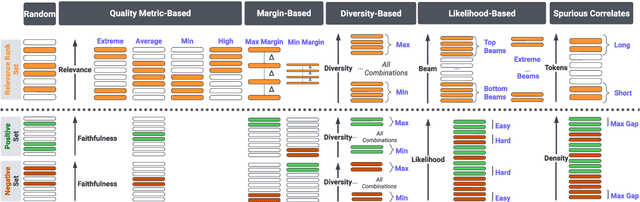
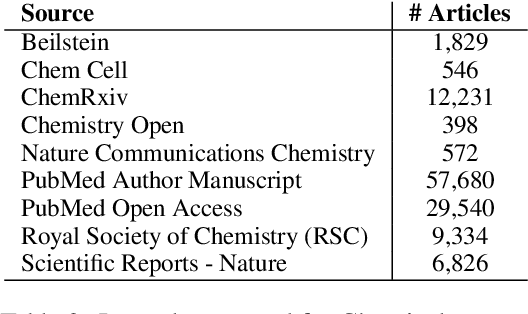
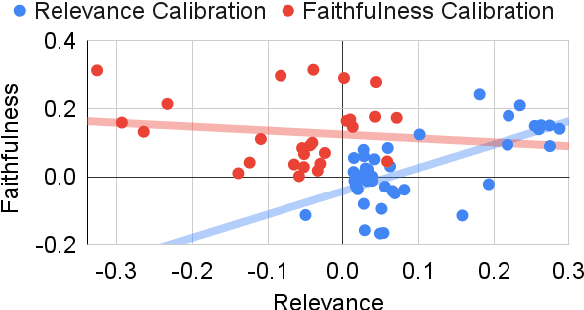
Abstract:Summarization models often generate text that is poorly calibrated to quality metrics because they are trained to maximize the likelihood of a single reference (MLE). To address this, recent work has added a calibration step, which exposes a model to its own ranked outputs to improve relevance or, in a separate line of work, contrasts positive and negative sets to improve faithfulness. While effective, much of this work has focused on how to generate and optimize these sets. Less is known about why one setup is more effective than another. In this work, we uncover the underlying characteristics of effective sets. For each training instance, we form a large, diverse pool of candidates and systematically vary the subsets used for calibration fine-tuning. Each selection strategy targets distinct aspects of the sets, such as lexical diversity or the size of the gap between positive and negatives. On three diverse scientific long-form summarization datasets (spanning biomedical, clinical, and chemical domains), we find, among others, that faithfulness calibration is optimal when the negative sets are extractive and more likely to be generated, whereas for relevance calibration, the metric margin between candidates should be maximized and surprise--the disagreement between model and metric defined candidate rankings--minimized. Code to create, select, and optimize calibration sets is available at https://github.com/griff4692/calibrating-summaries
A Bayesian Causal Inference Approach for Assessing Fairness in Clinical Decision-Making
Nov 21, 2022



Abstract:Fairness in clinical decision-making is a critical element of health equity, but assessing fairness of clinical decisions from observational data is challenging. Recently, many fairness notions have been proposed to quantify fairness in decision-making, among which causality-based fairness notions have gained increasing attention due to its potential in adjusting for confounding and reasoning about bias. However, causal fairness notions remain under-explored in the context of clinical decision-making with large-scale healthcare data. In this work, we propose a Bayesian causal inference approach for assessing a causal fairness notion called principal fairness in clinical settings. We demonstrate our approach using both simulated data and electronic health records (EHR) data.
Ontologizing Health Systems Data at Scale: Making Translational Discovery a Reality
Sep 10, 2022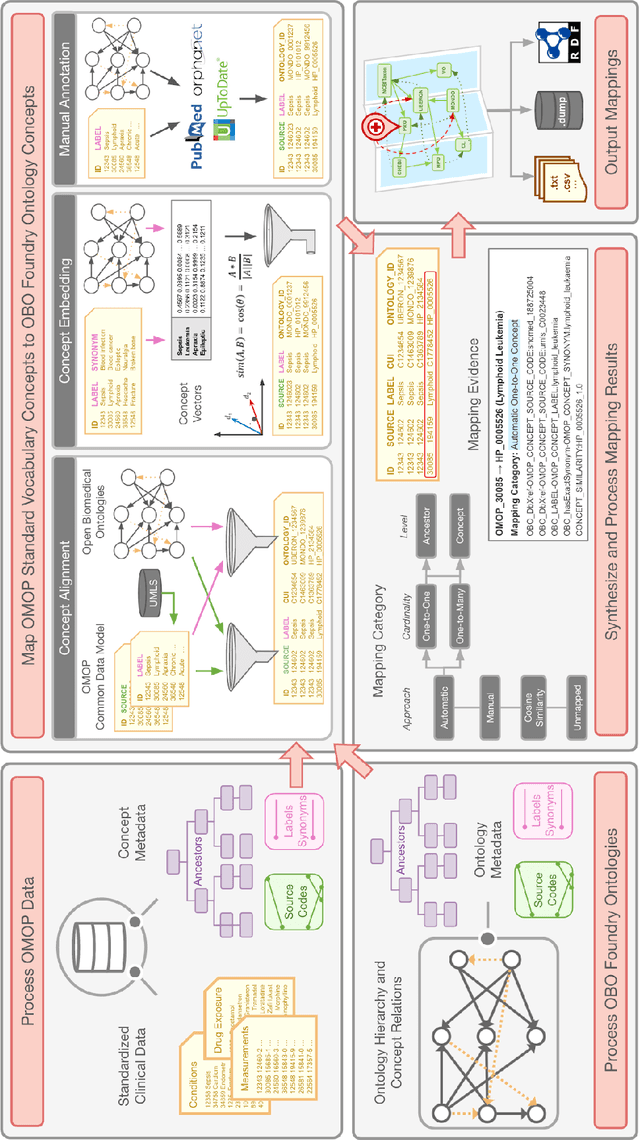
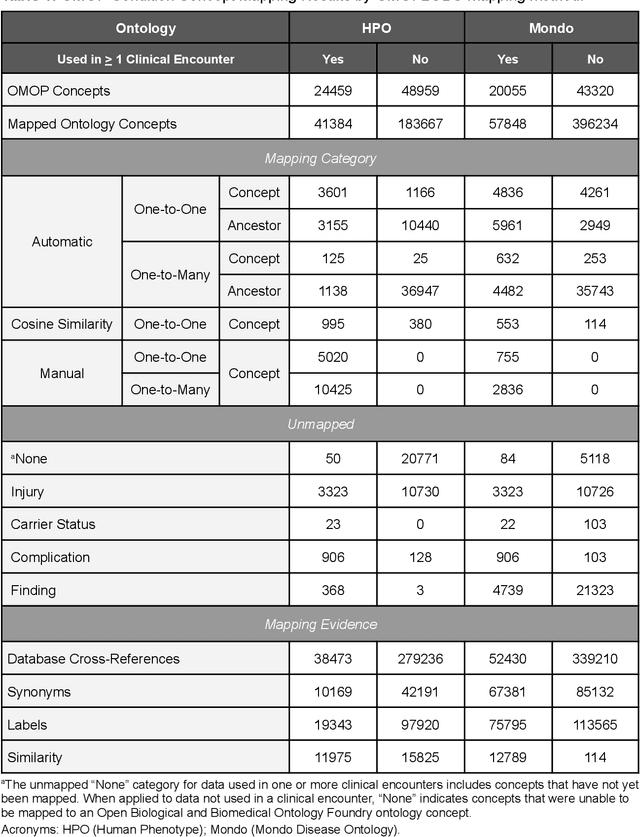
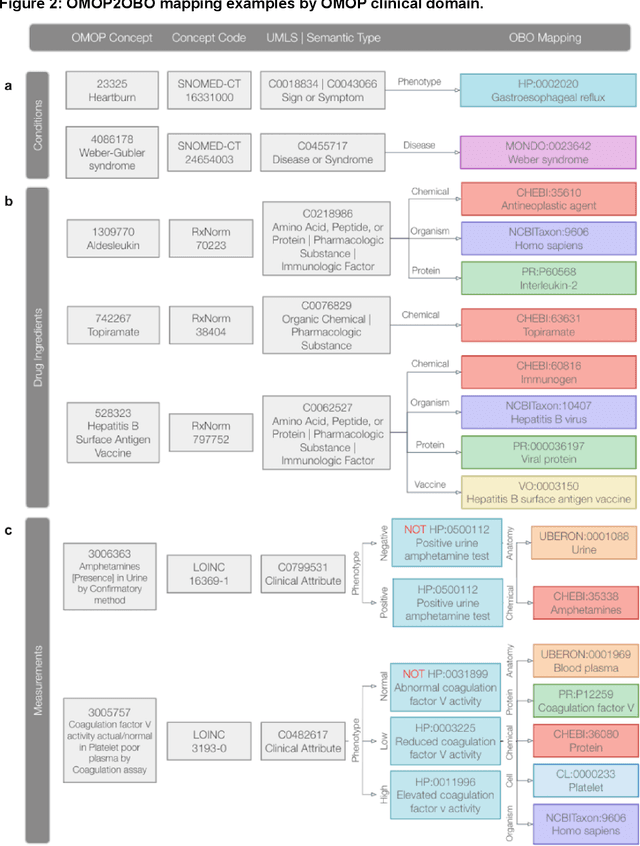
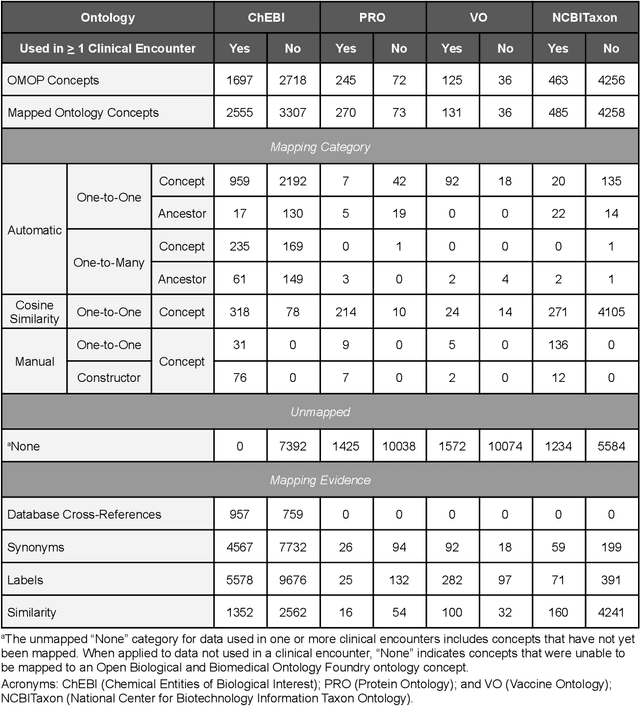
Abstract:Common data models solve many challenges of standardizing electronic health record (EHR) data, but are unable to semantically integrate the resources needed for deep phenotyping. Open Biological and Biomedical Ontology (OBO) Foundry ontologies provide semantically computable representations of biological knowledge and enable the integration of a variety of biomedical data. However, mapping EHR data to OBO Foundry ontologies requires significant manual curation and domain expertise. We introduce a framework for mapping Observational Medical Outcomes Partnership (OMOP) standard vocabularies to OBO Foundry ontologies. Using this framework, we produced mappings for 92,367 conditions, 8,615 drug ingredients, and 10,673 measurement results. Mapping accuracy was verified by domain experts and when examined across 24 hospitals, the mappings covered 99% of conditions and drug ingredients and 68% of measurements. Finally, we demonstrate that OMOP2OBO mappings can aid in the systematic identification of undiagnosed rare disease patients who might benefit from genetic testing.
The Medical Deconfounder: Assessing Treatment Effect with Electronic Health Records (EHRs)
Apr 03, 2019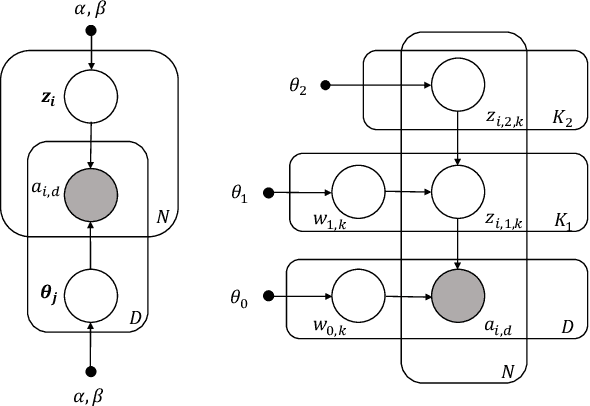
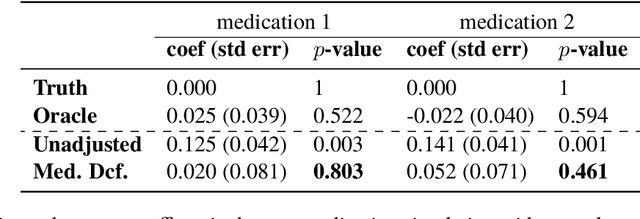
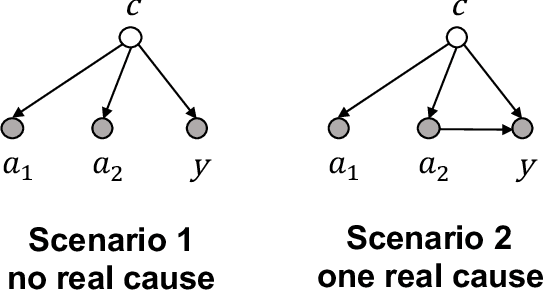
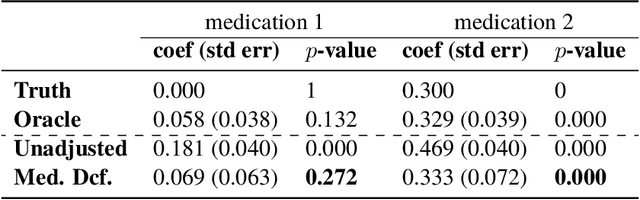
Abstract:Causal estimation of treatment effect has an important role in guiding physicians' decision process for drug prescription. While treatment effect is classically assessed with randomized controlled trials (RCTs), the availability of electronic health records (EHRs) bring an unprecedented opportunity for more efficient estimation. However, the presence of unobserved confounders makes treatment effect assessment from EHRs a challenging task. Confounders are the variables that affect both drug prescription and the patient's outcome; examples include a patient's gender, race, social economic status and comorbidities. When these confounders are unobserved, they bias the estimation. To adjust for unobserved confounders, we develop the medical deconfounder, a machine learning algorithm that unbiasedly estimates treatment effect from EHRs. The medical deconfounder first constructs a substitute confounder by modeling which drugs were prescribed to each patient; this substitute confounder is guaranteed to capture all multi-drug confounders, observed or unobserved (Wang and Blei, 2018). It then uses this substitute confounder to adjust for the confounding bias in the analysis. We validate the medical deconfounder on simulations and two medical data sets. The medical deconfounder produces closer-to-truth estimates in simulations and identifies effective medications that are more consistent with the findings reported in the medical literature compared to classical approaches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge