Richard D. Boyce
An Open-Source Knowledge Graph Ecosystem for the Life Sciences
Jul 11, 2023
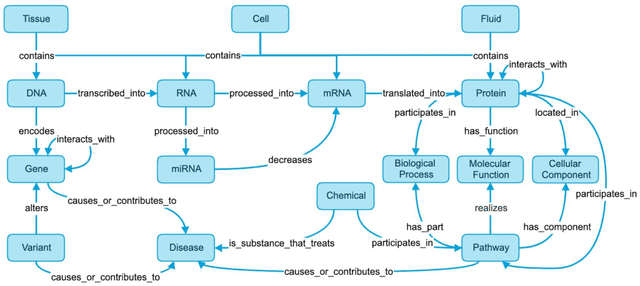
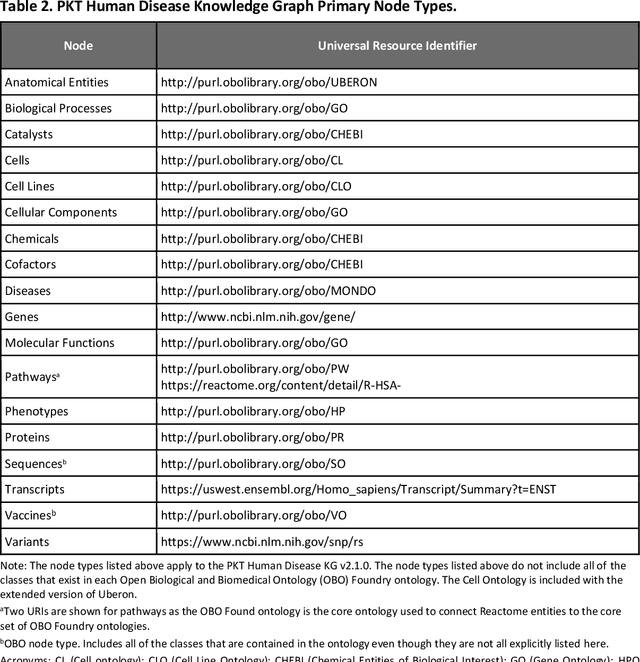
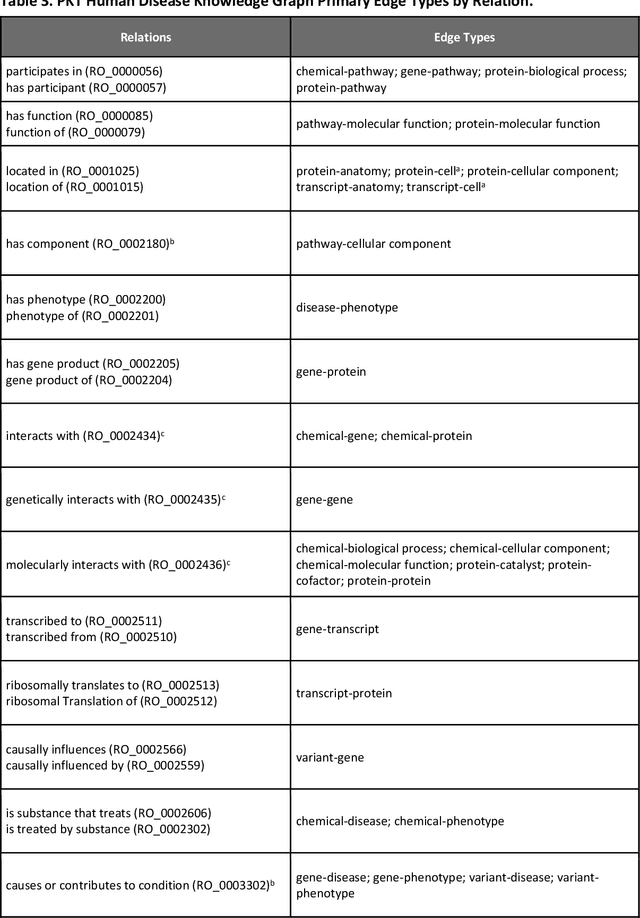
Abstract:Translational research requires data at multiple scales of biological organization. Advancements in sequencing and multi-omics technologies have increased the availability of these data but researchers face significant integration challenges. Knowledge graphs (KGs) are used to model complex phenomena, and methods exist to automatically construct them. However, tackling complex biomedical integration problems requires flexibility in the way knowledge is modeled. Moreover, existing KG construction methods provide robust tooling at the cost of fixed or limited choices among knowledge representation models. PheKnowLator (Phenotype Knowledge Translator) is a semantic ecosystem for automating the FAIR (Findable, Accessible, Interoperable, and Reusable) construction of ontologically grounded KGs with fully customizable knowledge representation. The ecosystem includes KG construction resources (e.g., data preparation APIs), analysis tools (e.g., SPARQL endpoints and abstraction algorithms), and benchmarks (e.g., prebuilt KGs and embeddings). We evaluate the ecosystem by surveying open-source KG construction methods and analyzing its computational performance when constructing 12 large-scale KGs. With flexible knowledge representation, PheKnowLator enables fully customizable KGs without compromising performance or usability.
Developing a Knowledge Graph Framework for Pharmacokinetic Natural Product-Drug Interactions
Sep 24, 2022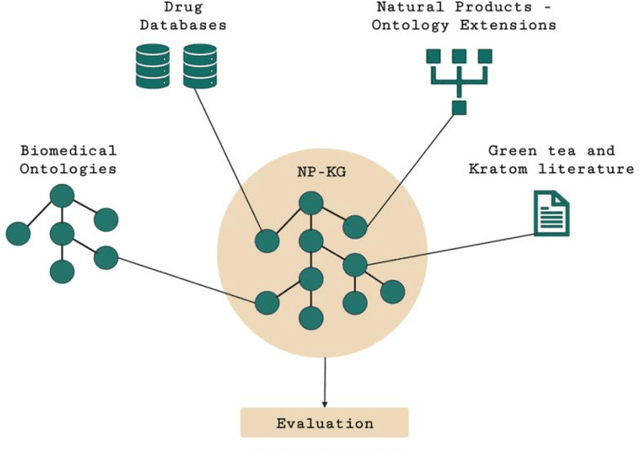
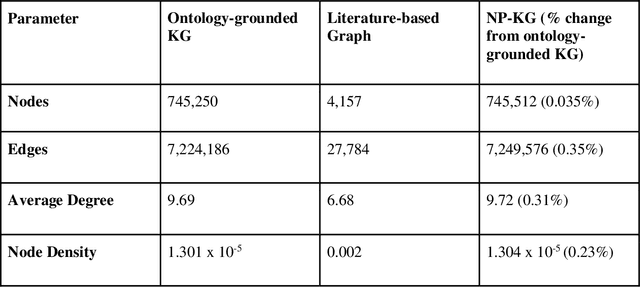
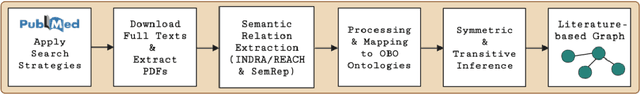
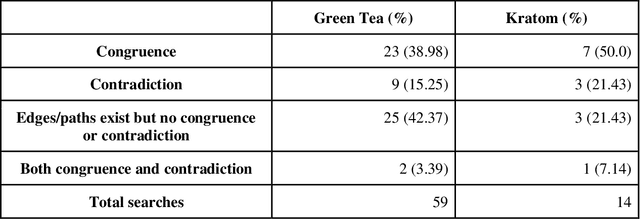
Abstract:Pharmacokinetic natural product-drug interactions (NPDIs) occur when botanical natural products are co-consumed with pharmaceutical drugs. Understanding mechanisms of NPDIs is key to preventing adverse events. We constructed a knowledge graph framework, NP-KG, as a step toward computational discovery of pharmacokinetic NPDIs. NP-KG is a heterogeneous KG with biomedical ontologies, linked data, and full texts of the scientific literature, constructed with the Phenotype Knowledge Translator framework and the semantic relation extraction systems, SemRep and Integrated Network and Dynamic Reasoning Assembler. NP-KG was evaluated with case studies of pharmacokinetic green tea- and kratom-drug interactions through path searches and meta-path discovery to determine congruent and contradictory information compared to ground truth data. The fully integrated NP-KG consisted of 745,512 nodes and 7,249,576 edges. Evaluation of NP-KG resulted in congruent (38.98% for green tea, 50% for kratom), contradictory (15.25% for green tea, 21.43% for kratom), and both congruent and contradictory (15.25% for green tea, 21.43% for kratom) information. Potential pharmacokinetic mechanisms for several purported NPDIs, including the green tea-raloxifene, green tea-nadolol, kratom-midazolam, kratom-quetiapine, and kratom-venlafaxine interactions were congruent with the published literature. NP-KG is the first KG to integrate biomedical ontologies with full texts of the scientific literature focused on natural products. We demonstrate the application of NP-KG to identify pharmacokinetic interactions involving enzymes, transporters, and pharmaceutical drugs. We envision that NP-KG will facilitate improved human-machine collaboration to guide researchers in future studies of pharmacokinetic NPDIs. The NP-KG framework is publicly available at https://doi.org/10.5281/zenodo.6814507 and https://github.com/sanyabt/np-kg.
Ontologizing Health Systems Data at Scale: Making Translational Discovery a Reality
Sep 10, 2022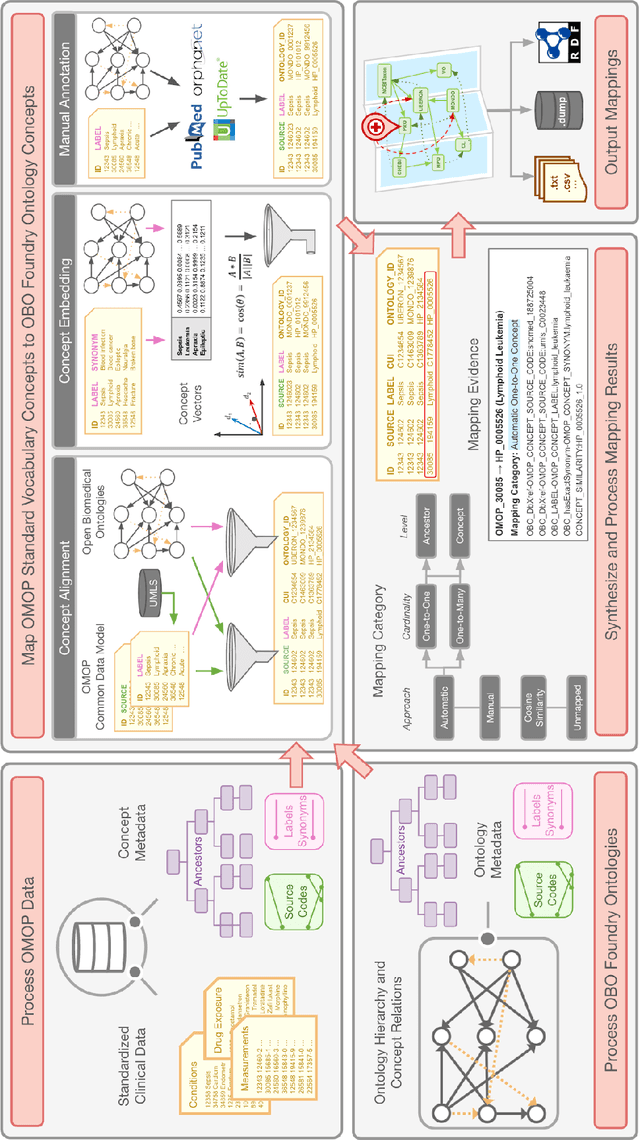
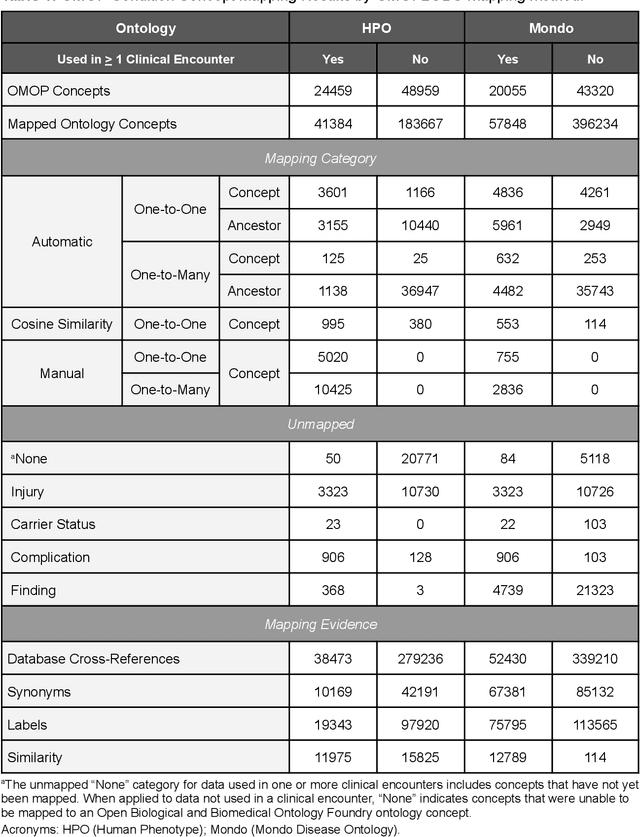
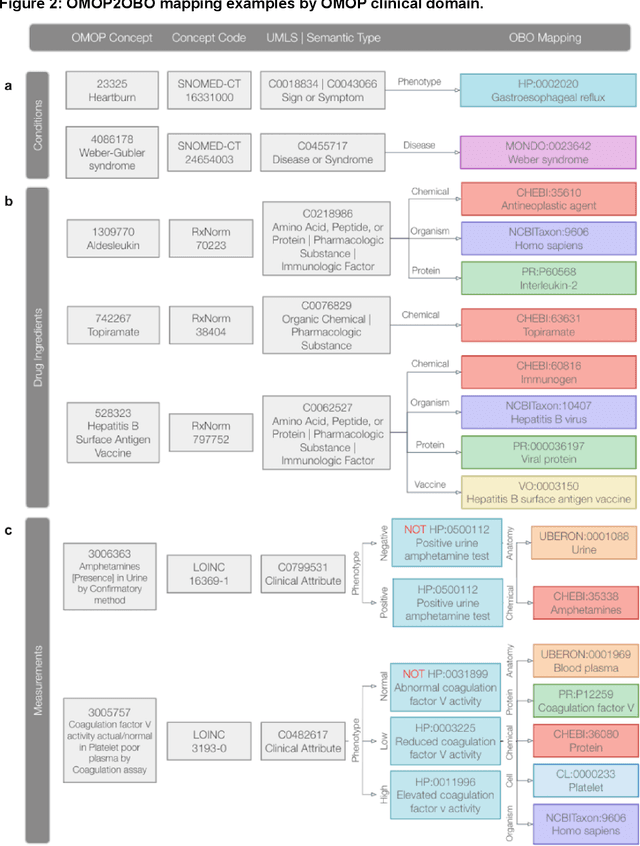
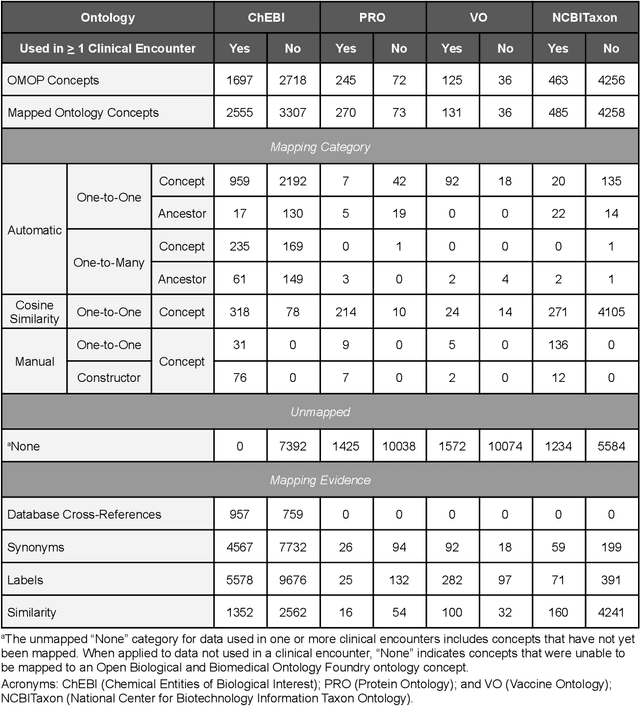
Abstract:Common data models solve many challenges of standardizing electronic health record (EHR) data, but are unable to semantically integrate the resources needed for deep phenotyping. Open Biological and Biomedical Ontology (OBO) Foundry ontologies provide semantically computable representations of biological knowledge and enable the integration of a variety of biomedical data. However, mapping EHR data to OBO Foundry ontologies requires significant manual curation and domain expertise. We introduce a framework for mapping Observational Medical Outcomes Partnership (OMOP) standard vocabularies to OBO Foundry ontologies. Using this framework, we produced mappings for 92,367 conditions, 8,615 drug ingredients, and 10,673 measurement results. Mapping accuracy was verified by domain experts and when examined across 24 hospitals, the mappings covered 99% of conditions and drug ingredients and 68% of measurements. Finally, we demonstrate that OMOP2OBO mappings can aid in the systematic identification of undiagnosed rare disease patients who might benefit from genetic testing.
Introducing Information Retrieval for Biomedical Informatics Students
May 06, 2021Abstract:Introducing biomedical informatics (BMI) students to natural language processing (NLP) requires balancing technical depth with practical know-how to address application-focused needs. We developed a set of three activities introducing introductory BMI students to information retrieval with NLP, covering document representation strategies and language models from TF-IDF to BERT. These activities provide students with hands-on experience targeted towards common use cases, and introduce fundamental components of NLP workflows for a wide variety of applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge