Denis Newman-Griffis
Disability data futures: Achievable imaginaries for AI and disability data justice
Nov 06, 2024Abstract:Data are the medium through which individuals' identities and experiences are filtered in contemporary states and systems, and AI is increasingly the layer mediating between people, data, and decisions. The history of data and AI is often one of disability exclusion, oppression, and the reduction of disabled experience; left unchallenged, the current proliferation of AI and data systems thus risks further automating ableism behind the veneer of algorithmic neutrality. However, exclusionary histories do not preclude inclusive futures, and disability-led visions can chart new paths for collective action to achieve futures founded in disability justice. This chapter brings together four academics and disability advocates working at the nexus of disability, data, and AI, to describe achievable imaginaries for artificial intelligence and disability data justice. Reflecting diverse contexts, disciplinary perspectives, and personal experiences, we draw out the shape, actors, and goals of imagined future systems where data and AI support movement towards disability justice.
Alternative models: Critical examination of disability definitions in the development of artificial intelligence technologies
Jun 16, 2022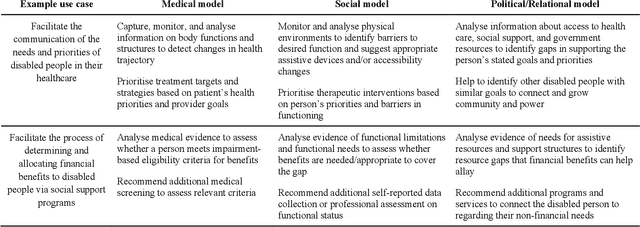
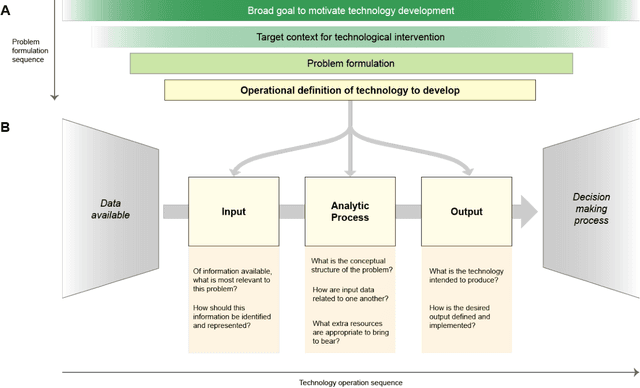
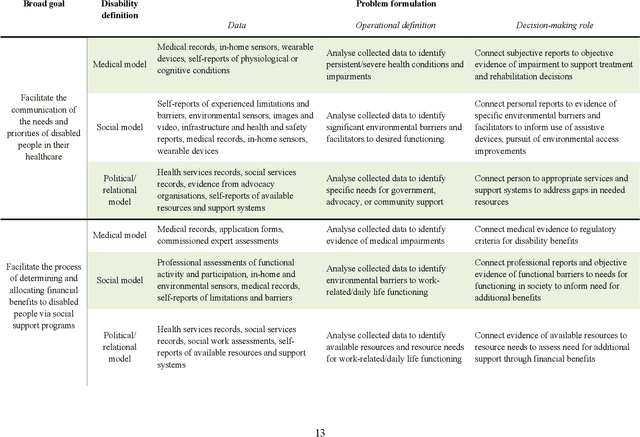
Abstract:Disabled people are subject to a wide variety of complex decision-making processes in diverse areas such as healthcare, employment, and government policy. These contexts, which are already often opaque to the people they affect and lack adequate representation of disabled perspectives, are rapidly adopting artificial intelligence (AI) technologies for data analytics to inform decision making, creating an increased risk of harm due to inappropriate or inequitable algorithms. This article presents a framework for critically examining AI data analytics technologies through a disability lens and investigates how the definition of disability chosen by the designers of an AI technology affects its impact on disabled subjects of analysis. We consider three conceptual models of disability: the medical model, the social model, and the relational model; and show how AI technologies designed under each of these models differ so significantly as to be incompatible with and contradictory to one another. Through a discussion of common use cases for AI analytics in healthcare and government disability benefits, we illustrate specific considerations and decision points in the technology design process that affect power dynamics and inclusion in these settings and help determine their orientation towards marginalisation or support. The framework we present can serve as a foundation for in-depth critical examination of AI technologies and the development of a design praxis for disability-related AI analytics.
Robust Knowledge Graph Completion with Stacked Convolutions and a Student Re-Ranking Network
Jun 11, 2021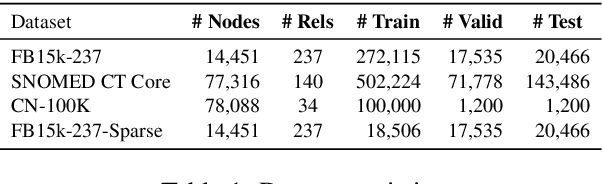
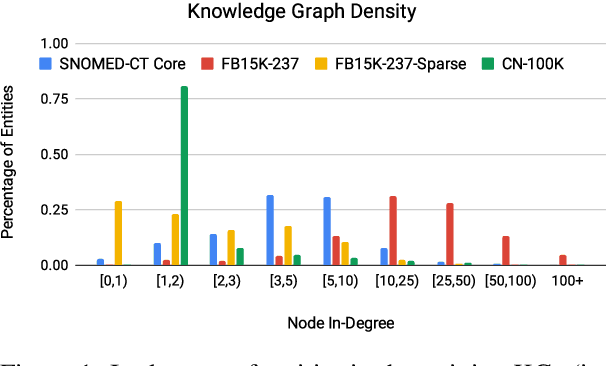
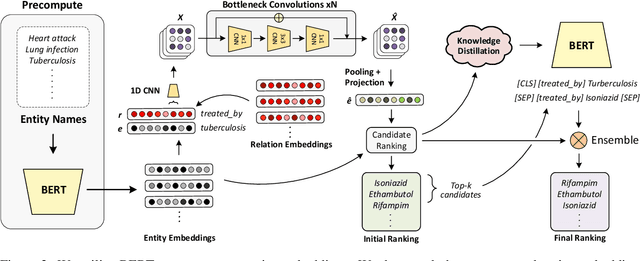

Abstract:Knowledge Graph (KG) completion research usually focuses on densely connected benchmark datasets that are not representative of real KGs. We curate two KG datasets that include biomedical and encyclopedic knowledge and use an existing commonsense KG dataset to explore KG completion in the more realistic setting where dense connectivity is not guaranteed. We develop a deep convolutional network that utilizes textual entity representations and demonstrate that our model outperforms recent KG completion methods in this challenging setting. We find that our model's performance improvements stem primarily from its robustness to sparsity. We then distill the knowledge from the convolutional network into a student network that re-ranks promising candidate entities. This re-ranking stage leads to further improvements in performance and demonstrates the effectiveness of entity re-ranking for KG completion.
Introducing Information Retrieval for Biomedical Informatics Students
May 06, 2021Abstract:Introducing biomedical informatics (BMI) students to natural language processing (NLP) requires balancing technical depth with practical know-how to address application-focused needs. We developed a set of three activities introducing introductory BMI students to information retrieval with NLP, covering document representation strategies and language models from TF-IDF to BERT. These activities provide students with hands-on experience targeted towards common use cases, and introduce fundamental components of NLP workflows for a wide variety of applications.
Translational NLP: A New Paradigm and General Principles for Natural Language Processing Research
Apr 16, 2021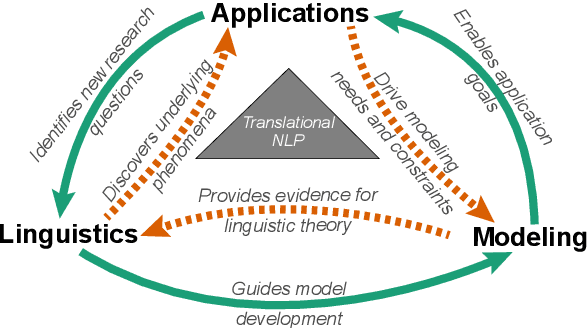
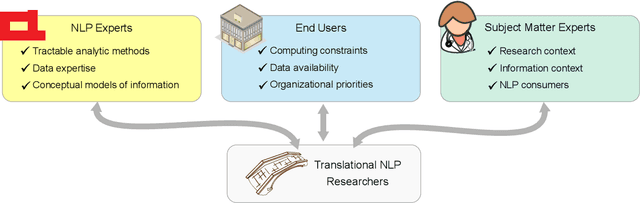
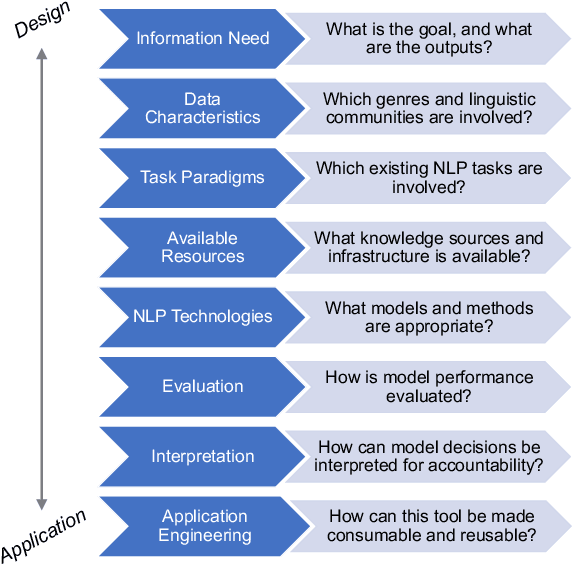
Abstract:Natural language processing (NLP) research combines the study of universal principles, through basic science, with applied science targeting specific use cases and settings. However, the process of exchange between basic NLP and applications is often assumed to emerge naturally, resulting in many innovations going unapplied and many important questions left unstudied. We describe a new paradigm of Translational NLP, which aims to structure and facilitate the processes by which basic and applied NLP research inform one another. Translational NLP thus presents a third research paradigm, focused on understanding the challenges posed by application needs and how these challenges can drive innovation in basic science and technology design. We show that many significant advances in NLP research have emerged from the intersection of basic principles with application needs, and present a conceptual framework outlining the stakeholders and key questions in translational research. Our framework provides a roadmap for developing Translational NLP as a dedicated research area, and identifies general translational principles to facilitate exchange between basic and applied research.
TextEssence: A Tool for Interactive Analysis of Semantic Shifts Between Corpora
Mar 19, 2021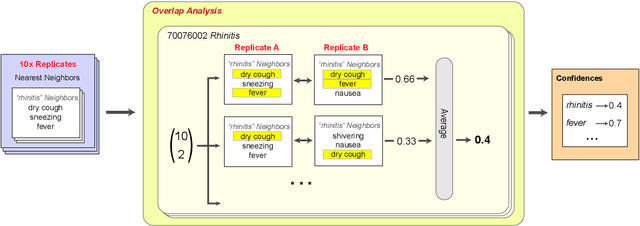
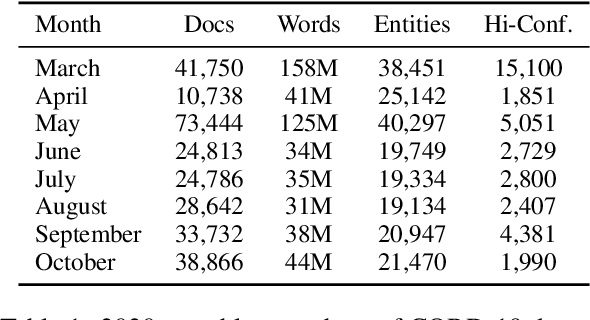
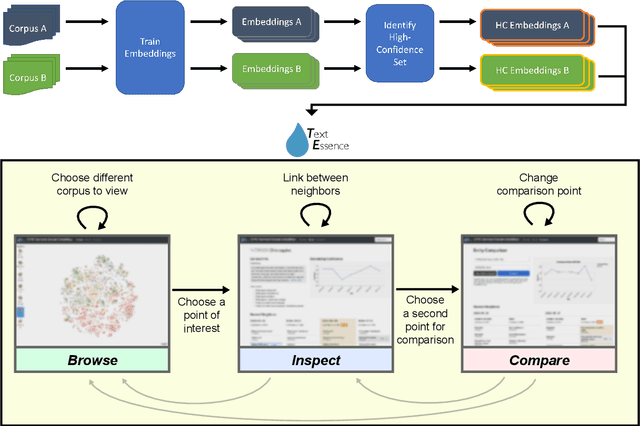
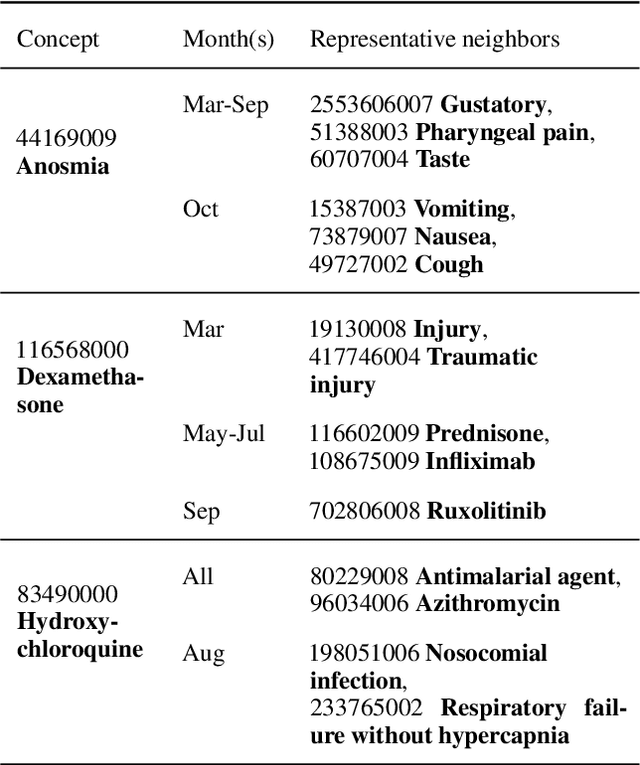
Abstract:Embeddings of words and concepts capture syntactic and semantic regularities of language; however, they have seen limited use as tools to study characteristics of different corpora and how they relate to one another. We introduce TextEssence, an interactive system designed to enable comparative analysis of corpora using embeddings. TextEssence includes visual, neighbor-based, and similarity-based modes of embedding analysis in a lightweight, web-based interface. We further propose a new measure of embedding confidence based on nearest neighborhood overlap, to assist in identifying high-quality embeddings for corpus analysis. A case study on COVID-19 scientific literature illustrates the utility of the system. TextEssence is available from https://github.com/drgriffis/text-essence.
Automated Coding of Under-Studied Medical Concept Domains: Linking Physical Activity Reports to the International Classification of Functioning, Disability, and Health
Nov 27, 2020
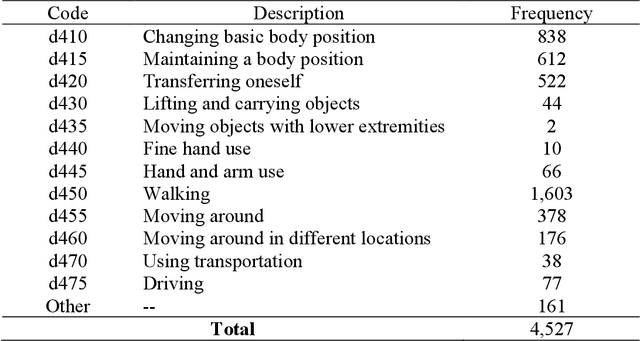
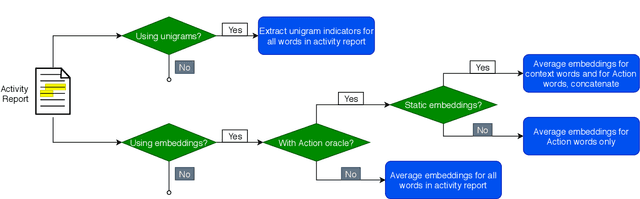
Abstract:Linking clinical narratives to standardized vocabularies and coding systems is a key component of unlocking the information in medical text for analysis. However, many domains of medical concepts lack well-developed terminologies that can support effective coding of medical text. We present a framework for developing natural language processing (NLP) technologies for automated coding of under-studied types of medical information, and demonstrate its applicability via a case study on physical mobility function. Mobility is a component of many health measures, from post-acute care and surgical outcomes to chronic frailty and disability, and is coded in the International Classification of Functioning, Disability, and Health (ICF). However, mobility and other types of functional activity remain under-studied in medical informatics, and neither the ICF nor commonly-used medical terminologies capture functional status terminology in practice. We investigated two data-driven paradigms, classification and candidate selection, to link narrative observations of mobility to standardized ICF codes, using a dataset of clinical narratives from physical therapy encounters. Recent advances in language modeling and word embedding were used as features for established machine learning models and a novel deep learning approach, achieving a macro F-1 score of 84% on linking mobility activity reports to ICF codes. Both classification and candidate selection approaches present distinct strengths for automated coding in under-studied domains, and we highlight that the combination of (i) a small annotated data set; (ii) expert definitions of codes of interest; and (iii) a representative text corpus is sufficient to produce high-performing automated coding systems. This study has implications for the ongoing growth of NLP tools for a variety of specialized applications in clinical care and research.
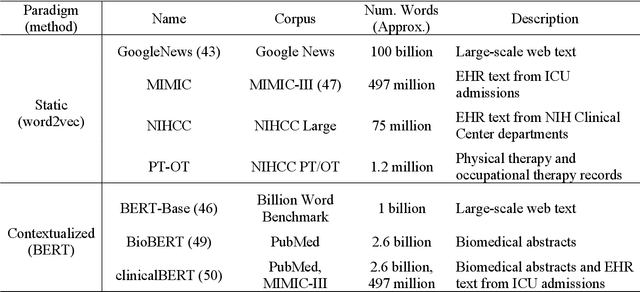
MedType: Improving Medical Entity Linking with Semantic Type Prediction
May 01, 2020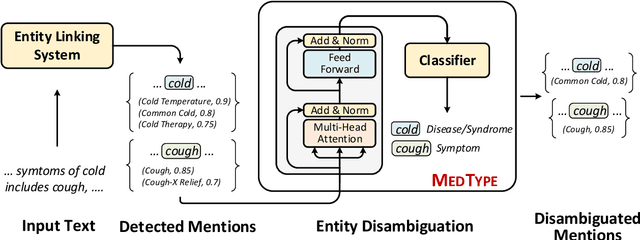
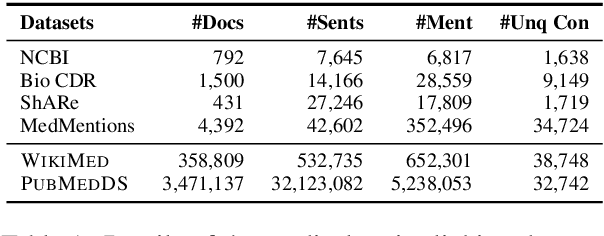
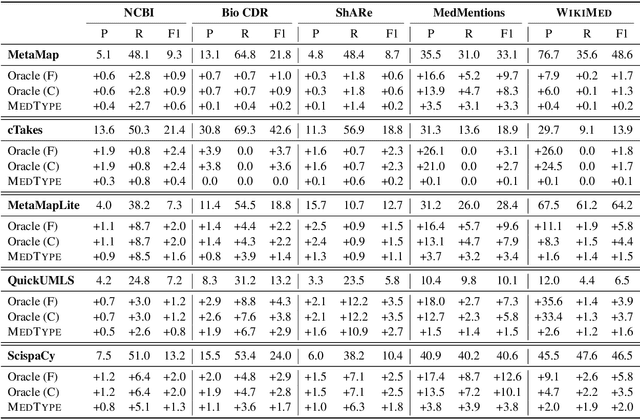
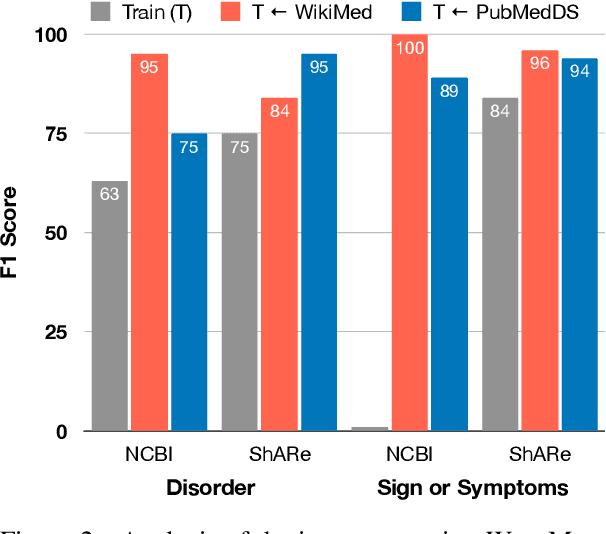
Abstract:Medical entity linking is the task of identifying and standardizing concepts referred in a scientific article or clinical record. Existing methods adopt a two-step approach of detecting mentions and identifying a list of candidate concepts for them. In this paper, we probe the impact of incorporating an entity disambiguation step in existing entity linkers. For this, we present MedType, a novel method that leverages the surrounding context to identify the semantic type of a mention and uses it for filtering out candidate concepts of the wrong types. We further present two novel largescale, automatically-created datasets of medical entity mentions: WIKIMED, a Wikipediabased dataset for cross-domain transfer learning, and PUBMEDDS, a distantly-supervised dataset of medical entity mentions in biomedical abstracts. Through extensive experiments across several datasets and methods, we demonstrate that MedType pre-trained on our proposed datasets substantially improve medical entity linking and gives state-of-the-art performance. We make our source code and datasets publicly available for medical entity linking research.
Writing habits and telltale neighbors: analyzing clinical concept usage patterns with sublanguage embeddings
Oct 01, 2019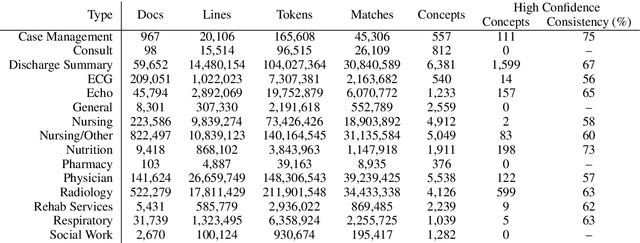
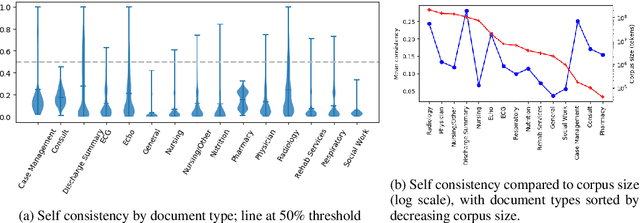
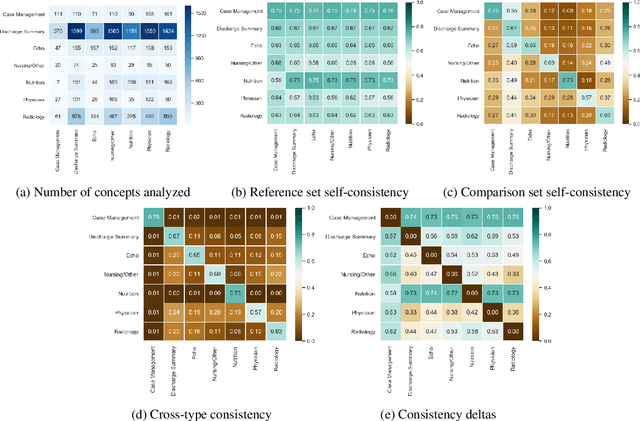
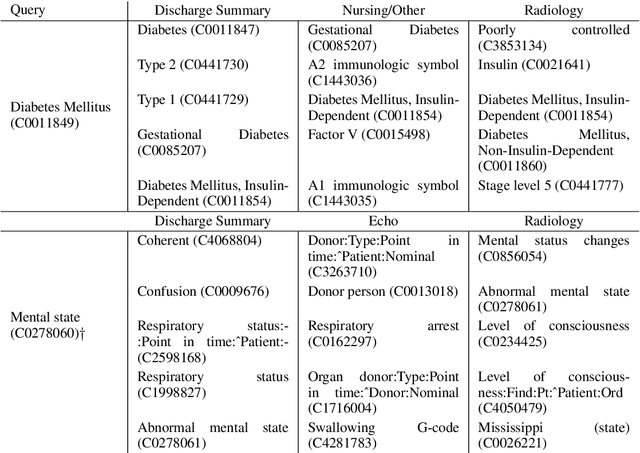
Abstract:Natural language processing techniques are being applied to increasingly diverse types of electronic health records, and can benefit from in-depth understanding of the distinguishing characteristics of medical document types. We present a method for characterizing the usage patterns of clinical concepts among different document types, in order to capture semantic differences beyond the lexical level. By training concept embeddings on clinical documents of different types and measuring the differences in their nearest neighborhood structures, we are able to measure divergences in concept usage while correcting for noise in embedding learning. Experiments on the MIMIC-III corpus demonstrate that our approach captures clinically-relevant differences in concept usage and provides an intuitive way to explore semantic characteristics of clinical document collections.
HARE: a Flexible Highlighting Annotator for Ranking and Exploration
Aug 29, 2019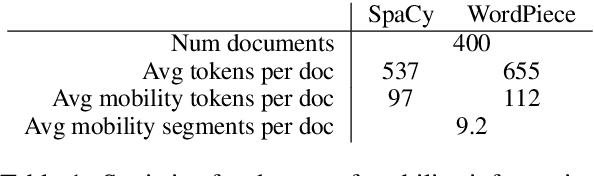
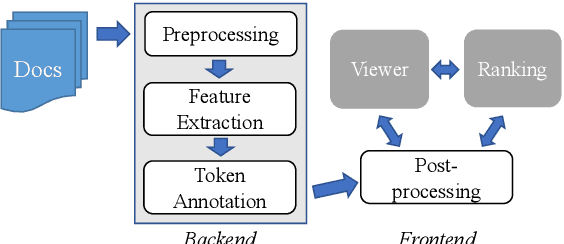
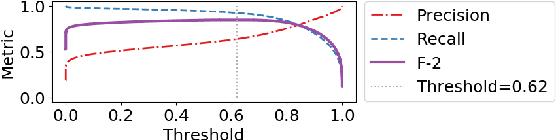
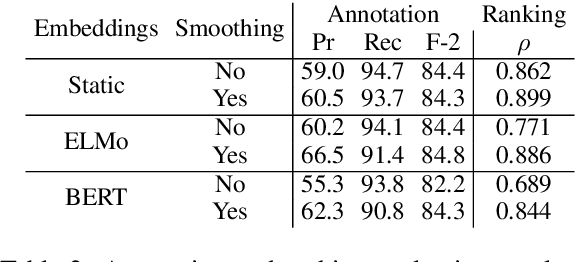
Abstract:Exploration and analysis of potential data sources is a significant challenge in the application of NLP techniques to novel information domains. We describe HARE, a system for highlighting relevant information in document collections to support ranking and triage, which provides tools for post-processing and qualitative analysis for model development and tuning. We apply HARE to the use case of narrative descriptions of mobility information in clinical data, and demonstrate its utility in comparing candidate embedding features. We provide a web-based interface for annotation visualization and document ranking, with a modular backend to support interoperability with existing annotation tools. Our system is available online at https://github.com/OSU-slatelab/HARE.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge