MedType: Improving Medical Entity Linking with Semantic Type Prediction
Paper and Code
May 01, 2020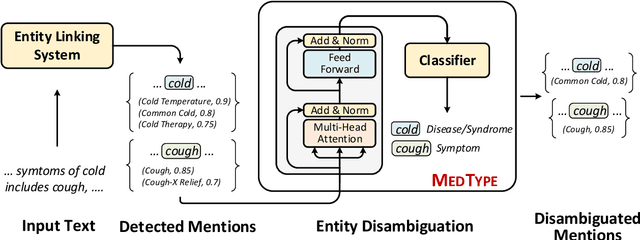
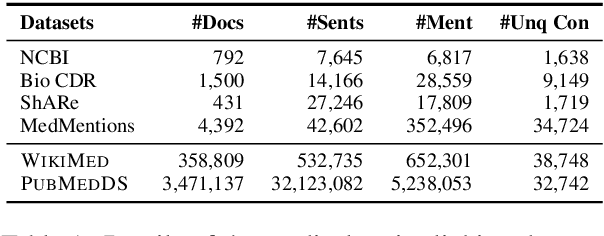
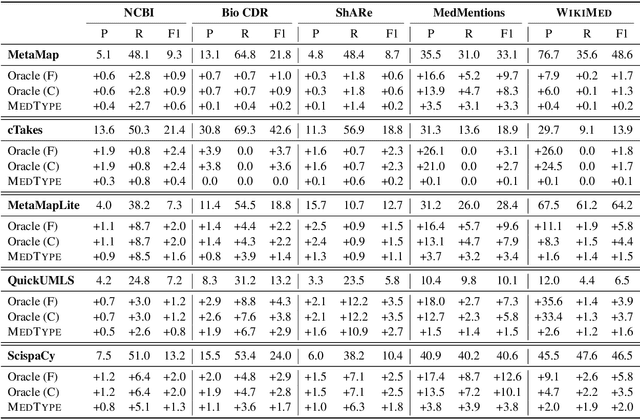
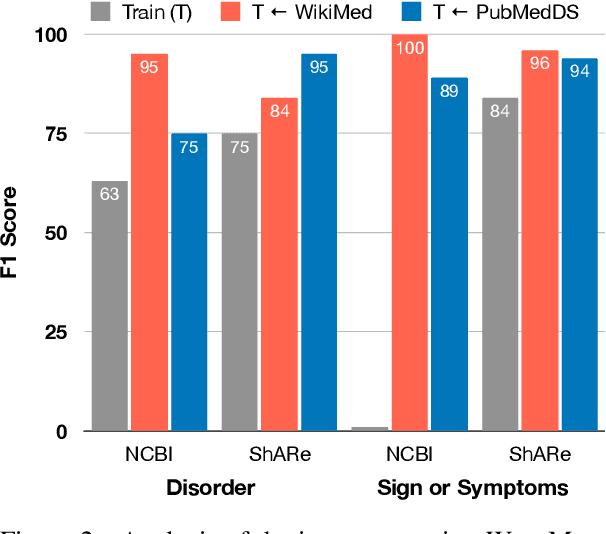
Medical entity linking is the task of identifying and standardizing concepts referred in a scientific article or clinical record. Existing methods adopt a two-step approach of detecting mentions and identifying a list of candidate concepts for them. In this paper, we probe the impact of incorporating an entity disambiguation step in existing entity linkers. For this, we present MedType, a novel method that leverages the surrounding context to identify the semantic type of a mention and uses it for filtering out candidate concepts of the wrong types. We further present two novel largescale, automatically-created datasets of medical entity mentions: WIKIMED, a Wikipediabased dataset for cross-domain transfer learning, and PUBMEDDS, a distantly-supervised dataset of medical entity mentions in biomedical abstracts. Through extensive experiments across several datasets and methods, we demonstrate that MedType pre-trained on our proposed datasets substantially improve medical entity linking and gives state-of-the-art performance. We make our source code and datasets publicly available for medical entity linking research.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge