Sandra L. Kane-Gill
Acute kidney injury prediction for non-critical care patients: a retrospective external and internal validation study
Feb 06, 2024Abstract:Background: Acute kidney injury (AKI), the decline of kidney excretory function, occurs in up to 18% of hospitalized admissions. Progression of AKI may lead to irreversible kidney damage. Methods: This retrospective cohort study includes adult patients admitted to a non-intensive care unit at the University of Pittsburgh Medical Center (UPMC) (n = 46,815) and University of Florida Health (UFH) (n = 127,202). We developed and compared deep learning and conventional machine learning models to predict progression to Stage 2 or higher AKI within the next 48 hours. We trained local models for each site (UFH Model trained on UFH, UPMC Model trained on UPMC) and a separate model with a development cohort of patients from both sites (UFH-UPMC Model). We internally and externally validated the models on each site and performed subgroup analyses across sex and race. Results: Stage 2 or higher AKI occurred in 3% (n=3,257) and 8% (n=2,296) of UFH and UPMC patients, respectively. Area under the receiver operating curve values (AUROC) for the UFH test cohort ranged between 0.77 (UPMC Model) and 0.81 (UFH Model), while AUROC values ranged between 0.79 (UFH Model) and 0.83 (UPMC Model) for the UPMC test cohort. UFH-UPMC Model achieved an AUROC of 0.81 (95% confidence interval [CI] [0.80, 0.83]) for UFH and 0.82 (95% CI [0.81,0.84]) for UPMC test cohorts; an area under the precision recall curve values (AUPRC) of 0.6 (95% CI, [0.05, 0.06]) for UFH and 0.13 (95% CI, [0.11,0.15]) for UPMC test cohorts. Kinetic estimated glomerular filtration rate, nephrotoxic drug burden and blood urea nitrogen remained the top three features with the highest influence across the models and health centers. Conclusion: Locally developed models displayed marginally reduced discrimination when tested on another institution, while the top set of influencing features remained the same across the models and sites.
Developing a Knowledge Graph Framework for Pharmacokinetic Natural Product-Drug Interactions
Sep 24, 2022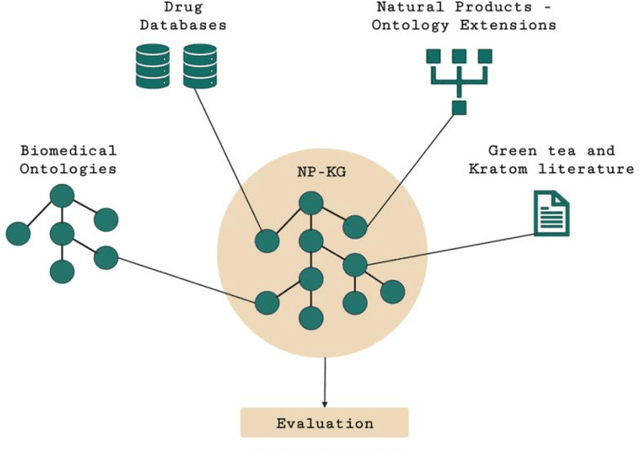
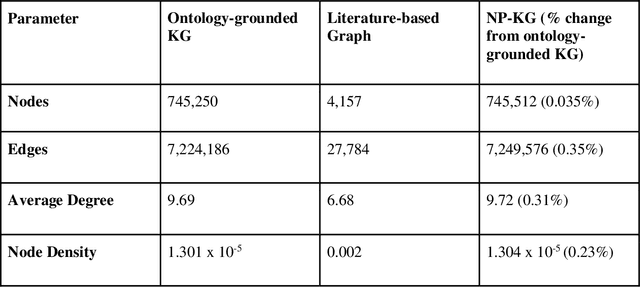
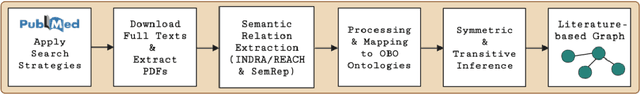
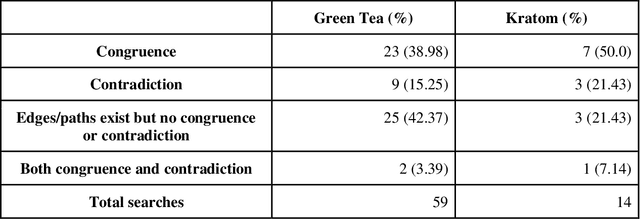
Abstract:Pharmacokinetic natural product-drug interactions (NPDIs) occur when botanical natural products are co-consumed with pharmaceutical drugs. Understanding mechanisms of NPDIs is key to preventing adverse events. We constructed a knowledge graph framework, NP-KG, as a step toward computational discovery of pharmacokinetic NPDIs. NP-KG is a heterogeneous KG with biomedical ontologies, linked data, and full texts of the scientific literature, constructed with the Phenotype Knowledge Translator framework and the semantic relation extraction systems, SemRep and Integrated Network and Dynamic Reasoning Assembler. NP-KG was evaluated with case studies of pharmacokinetic green tea- and kratom-drug interactions through path searches and meta-path discovery to determine congruent and contradictory information compared to ground truth data. The fully integrated NP-KG consisted of 745,512 nodes and 7,249,576 edges. Evaluation of NP-KG resulted in congruent (38.98% for green tea, 50% for kratom), contradictory (15.25% for green tea, 21.43% for kratom), and both congruent and contradictory (15.25% for green tea, 21.43% for kratom) information. Potential pharmacokinetic mechanisms for several purported NPDIs, including the green tea-raloxifene, green tea-nadolol, kratom-midazolam, kratom-quetiapine, and kratom-venlafaxine interactions were congruent with the published literature. NP-KG is the first KG to integrate biomedical ontologies with full texts of the scientific literature focused on natural products. We demonstrate the application of NP-KG to identify pharmacokinetic interactions involving enzymes, transporters, and pharmaceutical drugs. We envision that NP-KG will facilitate improved human-machine collaboration to guide researchers in future studies of pharmacokinetic NPDIs. The NP-KG framework is publicly available at https://doi.org/10.5281/zenodo.6814507 and https://github.com/sanyabt/np-kg.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge