Ziyuan Guan
Intelligent Critical Care Center, University of Florida, Gainesville, FL, Department of Medicine, College of Medicine, University of Florida, Gainesville, FL
MANDARIN: Mixture-of-Experts Framework for Dynamic Delirium and Coma Prediction in ICU Patients: Development and Validation of an Acute Brain Dysfunction Prediction Model
Mar 08, 2025Abstract:Acute brain dysfunction (ABD) is a common, severe ICU complication, presenting as delirium or coma and leading to prolonged stays, increased mortality, and cognitive decline. Traditional screening tools like the Glasgow Coma Scale (GCS), Confusion Assessment Method (CAM), and Richmond Agitation-Sedation Scale (RASS) rely on intermittent assessments, causing delays and inconsistencies. In this study, we propose MANDARIN (Mixture-of-Experts Framework for Dynamic Delirium and Coma Prediction in ICU Patients), a 1.5M-parameter mixture-of-experts neural network to predict ABD in real-time among ICU patients. The model integrates temporal and static data from the ICU to predict the brain status in the next 12 to 72 hours, using a multi-branch approach to account for current brain status. The MANDARIN model was trained on data from 92,734 patients (132,997 ICU admissions) from 2 hospitals between 2008-2019 and validated externally on data from 11,719 patients (14,519 ICU admissions) from 15 hospitals and prospectively on data from 304 patients (503 ICU admissions) from one hospital in 2021-2024. Three datasets were used: the University of Florida Health (UFH) dataset, the electronic ICU Collaborative Research Database (eICU), and the Medical Information Mart for Intensive Care (MIMIC)-IV dataset. MANDARIN significantly outperforms the baseline neurological assessment scores (GCS, CAM, and RASS) for delirium prediction in both external (AUROC 75.5% CI: 74.2%-76.8% vs 68.3% CI: 66.9%-69.5%) and prospective (AUROC 82.0% CI: 74.8%-89.2% vs 72.7% CI: 65.5%-81.0%) cohorts, as well as for coma prediction (external AUROC 87.3% CI: 85.9%-89.0% vs 72.8% CI: 70.6%-74.9%, and prospective AUROC 93.4% CI: 88.5%-97.9% vs 67.7% CI: 57.7%-76.8%) with a 12-hour lead time. This tool has the potential to assist clinicians in decision-making by continuously monitoring the brain status of patients in the ICU.
MANGO: Multimodal Acuity traNsformer for intelliGent ICU Outcomes
Dec 13, 2024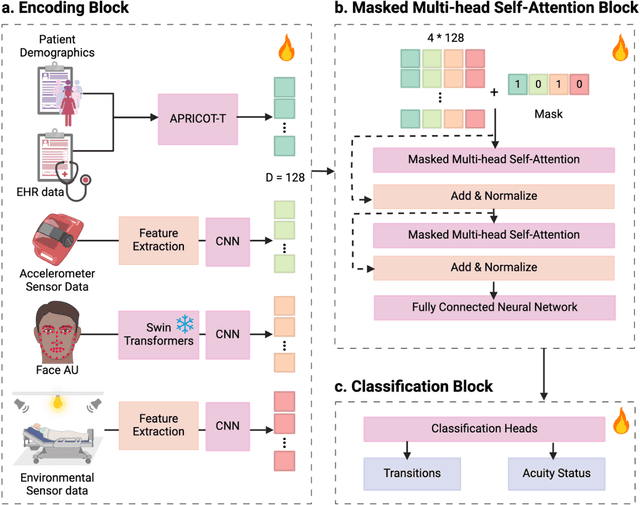
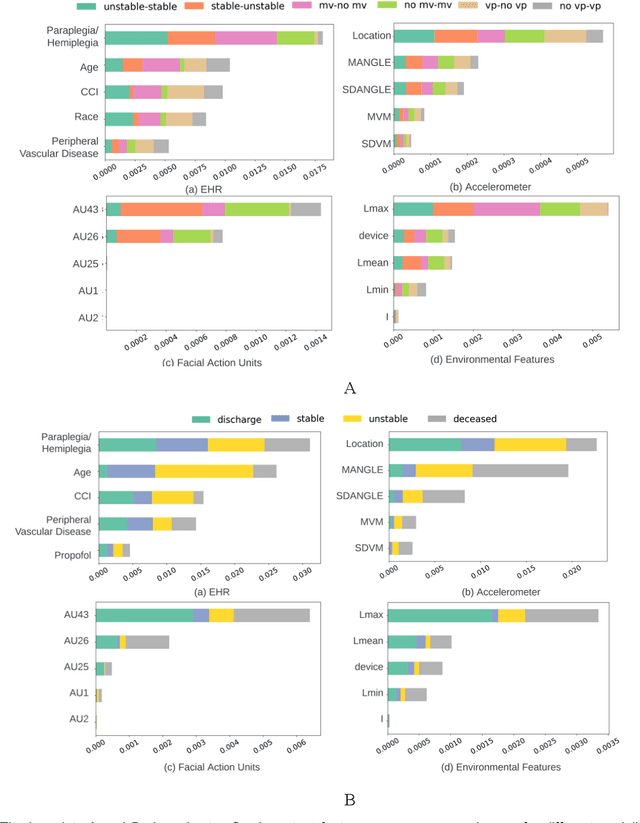
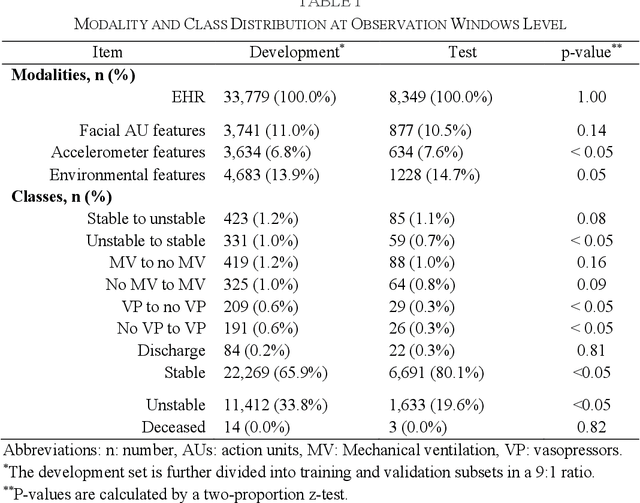
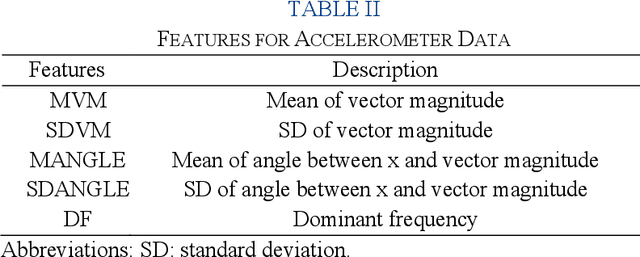
Abstract:Estimation of patient acuity in the Intensive Care Unit (ICU) is vital to ensure timely and appropriate interventions. Advances in artificial intelligence (AI) technologies have significantly improved the accuracy of acuity predictions. However, prior studies using machine learning for acuity prediction have predominantly relied on electronic health records (EHR) data, often overlooking other critical aspects of ICU stay, such as patient mobility, environmental factors, and facial cues indicating pain or agitation. To address this gap, we present MANGO: the Multimodal Acuity traNsformer for intelliGent ICU Outcomes, designed to enhance the prediction of patient acuity states, transitions, and the need for life-sustaining therapy. We collected a multimodal dataset ICU-Multimodal, incorporating four key modalities, EHR data, wearable sensor data, video of patient's facial cues, and ambient sensor data, which we utilized to train MANGO. The MANGO model employs a multimodal feature fusion network powered by Transformer masked self-attention method, enabling it to capture and learn complex interactions across these diverse data modalities even when some modalities are absent. Our results demonstrated that integrating multiple modalities significantly improved the model's ability to predict acuity status, transitions, and the need for life-sustaining therapy. The best-performing models achieved an area under the receiver operating characteristic curve (AUROC) of 0.76 (95% CI: 0.72-0.79) for predicting transitions in acuity status and the need for life-sustaining therapy, while 0.82 (95% CI: 0.69-0.89) for acuity status prediction...
DeLLiriuM: A large language model for delirium prediction in the ICU using structured EHR
Oct 22, 2024



Abstract:Delirium is an acute confusional state that has been shown to affect up to 31% of patients in the intensive care unit (ICU). Early detection of this condition could lead to more timely interventions and improved health outcomes. While artificial intelligence (AI) models have shown great potential for ICU delirium prediction using structured electronic health records (EHR), most of them have not explored the use of state-of-the-art AI models, have been limited to single hospitals, or have been developed and validated on small cohorts. The use of large language models (LLM), models with hundreds of millions to billions of parameters, with structured EHR data could potentially lead to improved predictive performance. In this study, we propose DeLLiriuM, a novel LLM-based delirium prediction model using EHR data available in the first 24 hours of ICU admission to predict the probability of a patient developing delirium during the rest of their ICU admission. We develop and validate DeLLiriuM on ICU admissions from 104,303 patients pertaining to 195 hospitals across three large databases: the eICU Collaborative Research Database, the Medical Information Mart for Intensive Care (MIMIC)-IV, and the University of Florida Health's Integrated Data Repository. The performance measured by the area under the receiver operating characteristic curve (AUROC) showed that DeLLiriuM outperformed all baselines in two external validation sets, with 0.77 (95% confidence interval 0.76-0.78) and 0.84 (95% confidence interval 0.83-0.85) across 77,543 patients spanning 194 hospitals. To the best of our knowledge, DeLLiriuM is the first LLM-based delirium prediction tool for the ICU based on structured EHR data, outperforming deep learning baselines which employ structured features and can provide helpful information to clinicians for timely interventions.
Transparent AI: Developing an Explainable Interface for Predicting Postoperative Complications
Apr 18, 2024



Abstract:Given the sheer volume of surgical procedures and the significant rate of postoperative fatalities, assessing and managing surgical complications has become a critical public health concern. Existing artificial intelligence (AI) tools for risk surveillance and diagnosis often lack adequate interpretability, fairness, and reproducibility. To address this, we proposed an Explainable AI (XAI) framework designed to answer five critical questions: why, why not, how, what if, and what else, with the goal of enhancing the explainability and transparency of AI models. We incorporated various techniques such as Local Interpretable Model-agnostic Explanations (LIME), SHapley Additive exPlanations (SHAP), counterfactual explanations, model cards, an interactive feature manipulation interface, and the identification of similar patients to address these questions. We showcased an XAI interface prototype that adheres to this framework for predicting major postoperative complications. This initial implementation has provided valuable insights into the vast explanatory potential of our XAI framework and represents an initial step towards its clinical adoption.
Global Contrastive Training for Multimodal Electronic Health Records with Language Supervision
Apr 10, 2024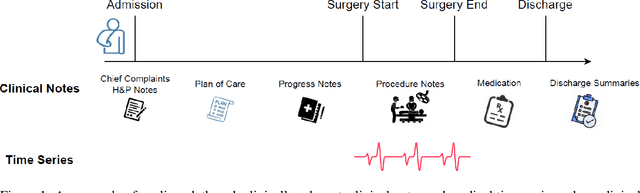
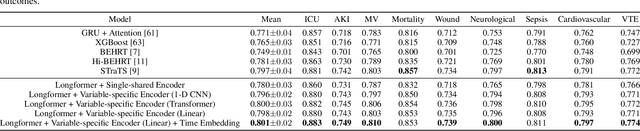
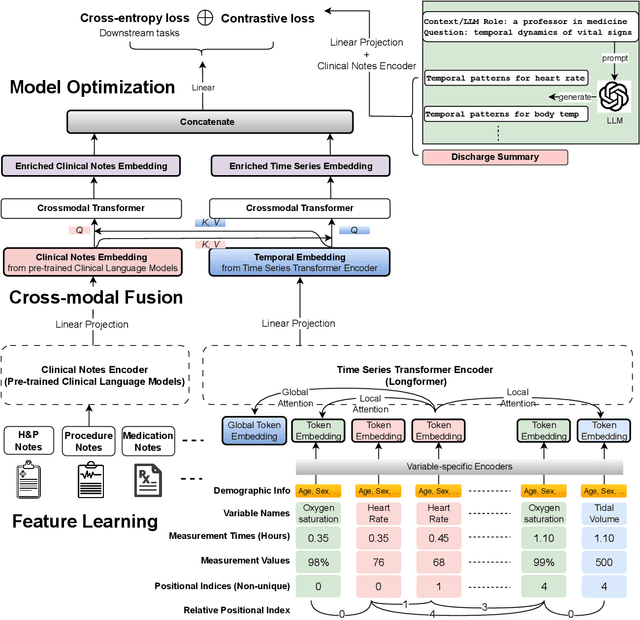
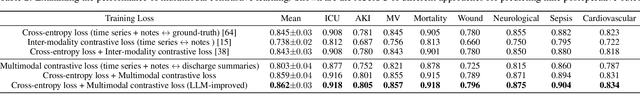
Abstract:Modern electronic health records (EHRs) hold immense promise in tracking personalized patient health trajectories through sequential deep learning, owing to their extensive breadth, scale, and temporal granularity. Nonetheless, how to effectively leverage multiple modalities from EHRs poses significant challenges, given its complex characteristics such as high dimensionality, multimodality, sparsity, varied recording frequencies, and temporal irregularities. To this end, this paper introduces a novel multimodal contrastive learning framework, specifically focusing on medical time series and clinical notes. To tackle the challenge of sparsity and irregular time intervals in medical time series, the framework integrates temporal cross-attention transformers with a dynamic embedding and tokenization scheme for learning multimodal feature representations. To harness the interconnected relationships between medical time series and clinical notes, the framework equips a global contrastive loss, aligning a patient's multimodal feature representations with the corresponding discharge summaries. Since discharge summaries uniquely pertain to individual patients and represent a holistic view of the patient's hospital stay, machine learning models are led to learn discriminative multimodal features via global contrasting. Extensive experiments with a real-world EHR dataset demonstrated that our framework outperformed state-of-the-art approaches on the exemplar task of predicting the occurrence of nine postoperative complications for more than 120,000 major inpatient surgeries using multimodal data from UF health system split among three hospitals (UF Health Gainesville, UF Health Jacksonville, and UF Health Jacksonville-North).
Federated learning model for predicting major postoperative complications
Apr 09, 2024
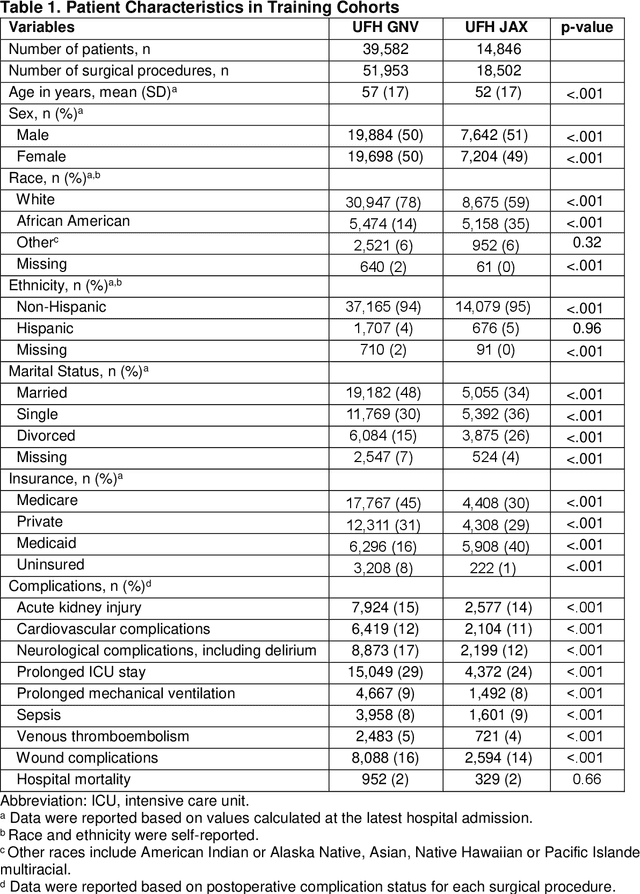


Abstract:Background: The accurate prediction of postoperative complication risk using Electronic Health Records (EHR) and artificial intelligence shows great potential. Training a robust artificial intelligence model typically requires large-scale and diverse datasets. In reality, collecting medical data often encounters challenges surrounding privacy protection. Methods: This retrospective cohort study includes adult patients who were admitted to UFH Gainesville (GNV) (n = 79,850) and Jacksonville (JAX) (n = 28,636) for any type of inpatient surgical procedure. Using perioperative and intraoperative features, we developed federated learning models to predict nine major postoperative complications (i.e., prolonged intensive care unit stay and mechanical ventilation). We compared federated learning models with local learning models trained on a single site and central learning models trained on pooled dataset from two centers. Results: Our federated learning models achieved the area under the receiver operating characteristics curve (AUROC) values ranged from 0.81 for wound complications to 0.92 for prolonged ICU stay at UFH GNV center. At UFH JAX center, these values ranged from 0.73-0.74 for wound complications to 0.92-0.93 for hospital mortality. Federated learning models achieved comparable AUROC performance to central learning models, except for prolonged ICU stay, where the performance of federated learning models was slightly higher than central learning models at UFH GNV center, but slightly lower at UFH JAX center. In addition, our federated learning model obtained comparable performance to the best local learning model at each center, demonstrating strong generalizability. Conclusion: Federated learning is shown to be a useful tool to train robust and generalizable models from large scale data across multiple institutions where data protection barriers are high.
A multi-cohort study on prediction of acute brain dysfunction states using selective state space models
Mar 11, 2024

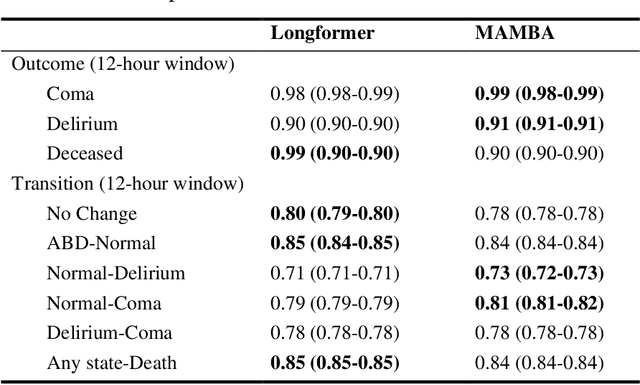

Abstract:Assessing acute brain dysfunction (ABD), including delirium and coma in the intensive care unit (ICU), is a critical challenge due to its prevalence and severe implications for patient outcomes. Current diagnostic methods rely on infrequent clinical observations, which can only determine a patient's ABD status after onset. Our research attempts to solve these problems by harnessing Electronic Health Records (EHR) data to develop automated methods for ABD prediction for patients in the ICU. Existing models solely predict a single state (e.g., either delirium or coma), require at least 24 hours of observation data to make predictions, do not dynamically predict fluctuating ABD conditions during ICU stay (typically a one-time prediction), and use small sample size, proprietary single-hospital datasets. Our research fills these gaps in the existing literature by dynamically predicting delirium, coma, and mortality for 12-hour intervals throughout an ICU stay and validating on two public datasets. Our research also introduces the concept of dynamically predicting critical transitions from non-ABD to ABD and between different ABD states in real time, which could be clinically more informative for the hospital staff. We compared the predictive performance of two state-of-the-art neural network models, the MAMBA selective state space model and the Longformer Transformer model. Using the MAMBA model, we achieved a mean area under the receiving operator characteristic curve (AUROC) of 0.95 on outcome prediction of ABD for 12-hour intervals. The model achieves a mean AUROC of 0.79 when predicting transitions between ABD states. Our study uses a curated dataset from the University of Florida Health Shands Hospital for internal validation and two publicly available datasets, MIMIC-IV and eICU, for external validation, demonstrating robustness across ICU stays from 203 hospitals and 140,945 patients.
Temporal Cross-Attention for Dynamic Embedding and Tokenization of Multimodal Electronic Health Records
Mar 06, 2024Abstract:The breadth, scale, and temporal granularity of modern electronic health records (EHR) systems offers great potential for estimating personalized and contextual patient health trajectories using sequential deep learning. However, learning useful representations of EHR data is challenging due to its high dimensionality, sparsity, multimodality, irregular and variable-specific recording frequency, and timestamp duplication when multiple measurements are recorded simultaneously. Although recent efforts to fuse structured EHR and unstructured clinical notes suggest the potential for more accurate prediction of clinical outcomes, less focus has been placed on EHR embedding approaches that directly address temporal EHR challenges by learning time-aware representations from multimodal patient time series. In this paper, we introduce a dynamic embedding and tokenization framework for precise representation of multimodal clinical time series that combines novel methods for encoding time and sequential position with temporal cross-attention. Our embedding and tokenization framework, when integrated into a multitask transformer classifier with sliding window attention, outperformed baseline approaches on the exemplar task of predicting the occurrence of nine postoperative complications of more than 120,000 major inpatient surgeries using multimodal data from three hospitals and two academic health centers in the United States.
Acute kidney injury prediction for non-critical care patients: a retrospective external and internal validation study
Feb 06, 2024Abstract:Background: Acute kidney injury (AKI), the decline of kidney excretory function, occurs in up to 18% of hospitalized admissions. Progression of AKI may lead to irreversible kidney damage. Methods: This retrospective cohort study includes adult patients admitted to a non-intensive care unit at the University of Pittsburgh Medical Center (UPMC) (n = 46,815) and University of Florida Health (UFH) (n = 127,202). We developed and compared deep learning and conventional machine learning models to predict progression to Stage 2 or higher AKI within the next 48 hours. We trained local models for each site (UFH Model trained on UFH, UPMC Model trained on UPMC) and a separate model with a development cohort of patients from both sites (UFH-UPMC Model). We internally and externally validated the models on each site and performed subgroup analyses across sex and race. Results: Stage 2 or higher AKI occurred in 3% (n=3,257) and 8% (n=2,296) of UFH and UPMC patients, respectively. Area under the receiver operating curve values (AUROC) for the UFH test cohort ranged between 0.77 (UPMC Model) and 0.81 (UFH Model), while AUROC values ranged between 0.79 (UFH Model) and 0.83 (UPMC Model) for the UPMC test cohort. UFH-UPMC Model achieved an AUROC of 0.81 (95% confidence interval [CI] [0.80, 0.83]) for UFH and 0.82 (95% CI [0.81,0.84]) for UPMC test cohorts; an area under the precision recall curve values (AUPRC) of 0.6 (95% CI, [0.05, 0.06]) for UFH and 0.13 (95% CI, [0.11,0.15]) for UPMC test cohorts. Kinetic estimated glomerular filtration rate, nephrotoxic drug burden and blood urea nitrogen remained the top three features with the highest influence across the models and health centers. Conclusion: Locally developed models displayed marginally reduced discrimination when tested on another institution, while the top set of influencing features remained the same across the models and sites.
APRICOT: Acuity Prediction in Intensive Care Unit (ICU): Predicting Stability, Transitions, and Life-Sustaining Therapies
Nov 03, 2023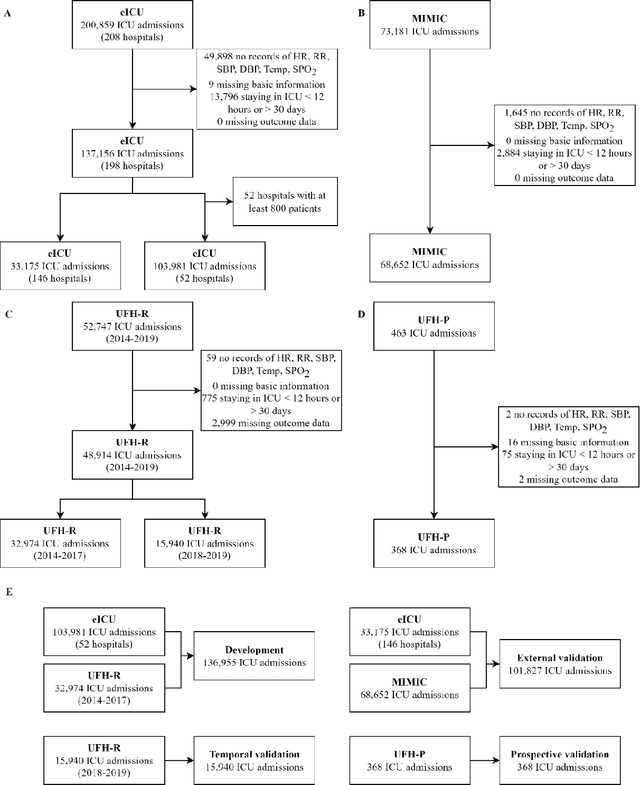

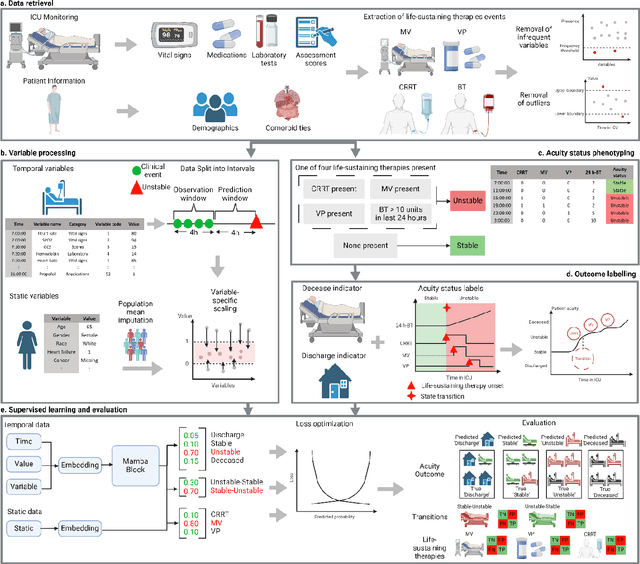
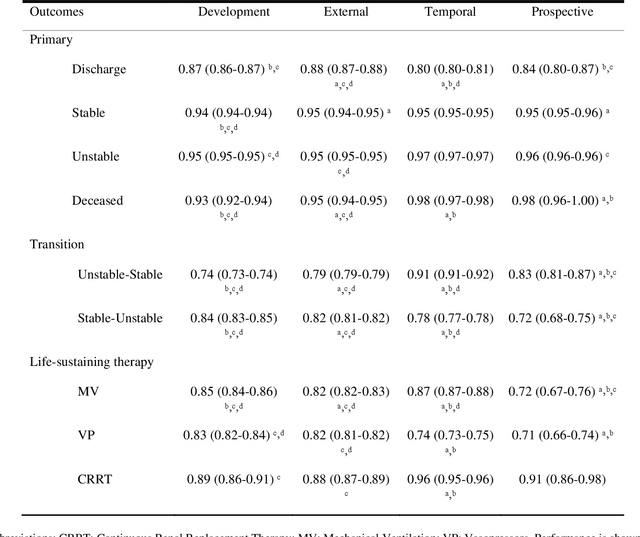
Abstract:The acuity state of patients in the intensive care unit (ICU) can quickly change from stable to unstable, sometimes leading to life-threatening conditions. Early detection of deteriorating conditions can result in providing more timely interventions and improved survival rates. Current approaches rely on manual daily assessments. Some data-driven approaches have been developed, that use mortality as a proxy of acuity in the ICU. However, these methods do not integrate acuity states to determine the stability of a patient or the need for life-sustaining therapies. In this study, we propose APRICOT (Acuity Prediction in Intensive Care Unit), a Transformer-based neural network to predict acuity state in real-time in ICU patients. We develop and extensively validate externally, temporally, and prospectively the APRICOT model on three large datasets: University of Florida Health (UFH), eICU Collaborative Research Database (eICU), and Medical Information Mart for Intensive Care (MIMIC)-IV. The performance of APRICOT shows comparable results to state-of-the-art mortality prediction models (external AUROC 0.93-0.93, temporal AUROC 0.96-0.98, and prospective AUROC 0.98) as well as acuity prediction models (external AUROC 0.80-0.81, temporal AUROC 0.77-0.78, and prospective AUROC 0.87). Furthermore, APRICOT can make predictions for the need for life-sustaining therapies, showing comparable results to state-of-the-art ventilation prediction models (external AUROC 0.80-0.81, temporal AUROC 0.87-0.88, and prospective AUROC 0.85), and vasopressor prediction models (external AUROC 0.82-0.83, temporal AUROC 0.73-0.75, prospective AUROC 0.87). This tool allows for real-time acuity monitoring of a patient and can provide helpful information to clinicians to make timely interventions. Furthermore, the model can suggest life-sustaining therapies that the patient might need in the next hours in the ICU.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge