Yingbo Ma
Transparent AI: Developing an Explainable Interface for Predicting Postoperative Complications
Apr 18, 2024



Abstract:Given the sheer volume of surgical procedures and the significant rate of postoperative fatalities, assessing and managing surgical complications has become a critical public health concern. Existing artificial intelligence (AI) tools for risk surveillance and diagnosis often lack adequate interpretability, fairness, and reproducibility. To address this, we proposed an Explainable AI (XAI) framework designed to answer five critical questions: why, why not, how, what if, and what else, with the goal of enhancing the explainability and transparency of AI models. We incorporated various techniques such as Local Interpretable Model-agnostic Explanations (LIME), SHapley Additive exPlanations (SHAP), counterfactual explanations, model cards, an interactive feature manipulation interface, and the identification of similar patients to address these questions. We showcased an XAI interface prototype that adheres to this framework for predicting major postoperative complications. This initial implementation has provided valuable insights into the vast explanatory potential of our XAI framework and represents an initial step towards its clinical adoption.
Global Contrastive Training for Multimodal Electronic Health Records with Language Supervision
Apr 10, 2024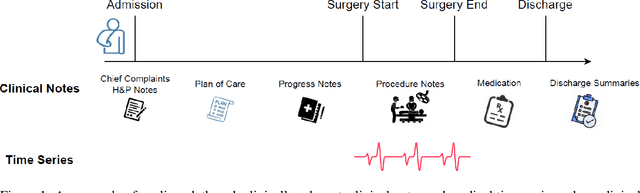
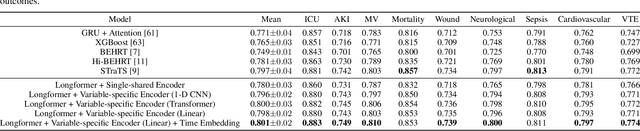
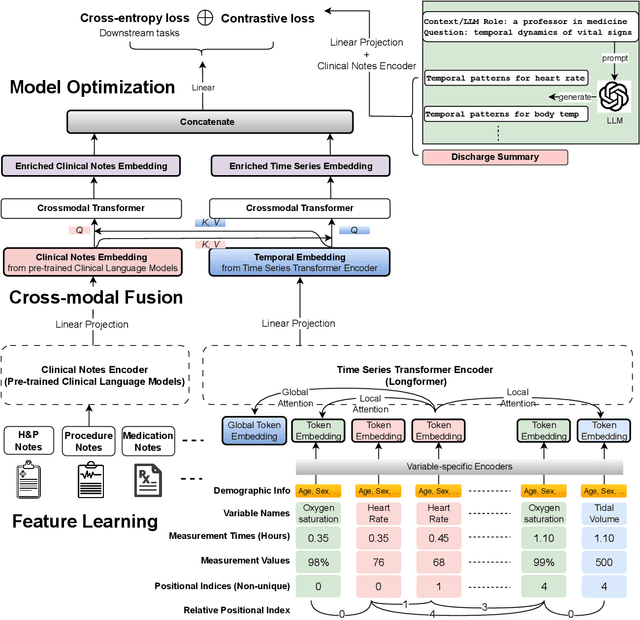
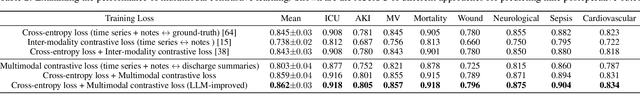
Abstract:Modern electronic health records (EHRs) hold immense promise in tracking personalized patient health trajectories through sequential deep learning, owing to their extensive breadth, scale, and temporal granularity. Nonetheless, how to effectively leverage multiple modalities from EHRs poses significant challenges, given its complex characteristics such as high dimensionality, multimodality, sparsity, varied recording frequencies, and temporal irregularities. To this end, this paper introduces a novel multimodal contrastive learning framework, specifically focusing on medical time series and clinical notes. To tackle the challenge of sparsity and irregular time intervals in medical time series, the framework integrates temporal cross-attention transformers with a dynamic embedding and tokenization scheme for learning multimodal feature representations. To harness the interconnected relationships between medical time series and clinical notes, the framework equips a global contrastive loss, aligning a patient's multimodal feature representations with the corresponding discharge summaries. Since discharge summaries uniquely pertain to individual patients and represent a holistic view of the patient's hospital stay, machine learning models are led to learn discriminative multimodal features via global contrasting. Extensive experiments with a real-world EHR dataset demonstrated that our framework outperformed state-of-the-art approaches on the exemplar task of predicting the occurrence of nine postoperative complications for more than 120,000 major inpatient surgeries using multimodal data from UF health system split among three hospitals (UF Health Gainesville, UF Health Jacksonville, and UF Health Jacksonville-North).
Federated learning model for predicting major postoperative complications
Apr 09, 2024
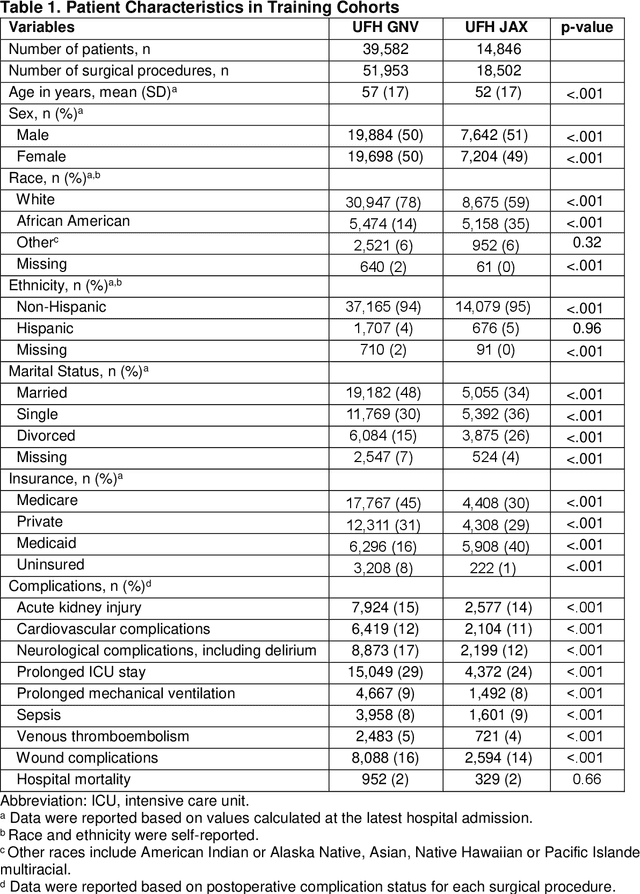


Abstract:Background: The accurate prediction of postoperative complication risk using Electronic Health Records (EHR) and artificial intelligence shows great potential. Training a robust artificial intelligence model typically requires large-scale and diverse datasets. In reality, collecting medical data often encounters challenges surrounding privacy protection. Methods: This retrospective cohort study includes adult patients who were admitted to UFH Gainesville (GNV) (n = 79,850) and Jacksonville (JAX) (n = 28,636) for any type of inpatient surgical procedure. Using perioperative and intraoperative features, we developed federated learning models to predict nine major postoperative complications (i.e., prolonged intensive care unit stay and mechanical ventilation). We compared federated learning models with local learning models trained on a single site and central learning models trained on pooled dataset from two centers. Results: Our federated learning models achieved the area under the receiver operating characteristics curve (AUROC) values ranged from 0.81 for wound complications to 0.92 for prolonged ICU stay at UFH GNV center. At UFH JAX center, these values ranged from 0.73-0.74 for wound complications to 0.92-0.93 for hospital mortality. Federated learning models achieved comparable AUROC performance to central learning models, except for prolonged ICU stay, where the performance of federated learning models was slightly higher than central learning models at UFH GNV center, but slightly lower at UFH JAX center. In addition, our federated learning model obtained comparable performance to the best local learning model at each center, demonstrating strong generalizability. Conclusion: Federated learning is shown to be a useful tool to train robust and generalizable models from large scale data across multiple institutions where data protection barriers are high.
Temporal Cross-Attention for Dynamic Embedding and Tokenization of Multimodal Electronic Health Records
Mar 06, 2024Abstract:The breadth, scale, and temporal granularity of modern electronic health records (EHR) systems offers great potential for estimating personalized and contextual patient health trajectories using sequential deep learning. However, learning useful representations of EHR data is challenging due to its high dimensionality, sparsity, multimodality, irregular and variable-specific recording frequency, and timestamp duplication when multiple measurements are recorded simultaneously. Although recent efforts to fuse structured EHR and unstructured clinical notes suggest the potential for more accurate prediction of clinical outcomes, less focus has been placed on EHR embedding approaches that directly address temporal EHR challenges by learning time-aware representations from multimodal patient time series. In this paper, we introduce a dynamic embedding and tokenization framework for precise representation of multimodal clinical time series that combines novel methods for encoding time and sequential position with temporal cross-attention. Our embedding and tokenization framework, when integrated into a multitask transformer classifier with sliding window attention, outperformed baseline approaches on the exemplar task of predicting the occurrence of nine postoperative complications of more than 120,000 major inpatient surgeries using multimodal data from three hospitals and two academic health centers in the United States.
High-performance symbolic-numerics via multiple dispatch
May 12, 2021
Abstract:As mathematical computing becomes more democratized in high-level languages, high-performance symbolic-numeric systems are necessary for domain scientists and engineers to get the best performance out of their machine without deep knowledge of code optimization. Naturally, users need different term types either to have different algebraic properties for them, or to use efficient data structures. To this end, we developed Symbolics.jl, an extendable symbolic system which uses dynamic multiple dispatch to change behavior depending on the domain needs. In this work we detail an underlying abstract term interface which allows for speed without sacrificing generality. We show that by formalizing a generic API on actions independent of implementation, we can retroactively add optimized data structures to our system without changing the pre-existing term rewriters. We showcase how this can be used to optimize term construction and give a 113x acceleration on general symbolic transformations. Further, we show that such a generic API allows for complementary term-rewriting implementations. We demonstrate the ability to swap between classical term-rewriting simplifiers and e-graph-based term-rewriting simplifiers. We showcase an e-graph ruleset which minimizes the number of CPU cycles during expression evaluation, and demonstrate how it simplifies a real-world reaction-network simulation to halve the runtime. Additionally, we show a reaction-diffusion partial differential equation solver which is able to be automatically converted into symbolic expressions via multiple dispatch tracing, which is subsequently accelerated and parallelized to give a 157x simulation speedup. Together, this presents Symbolics.jl as a next-generation symbolic-numeric computing environment geared towards modeling and simulation.
Opening the Blackbox: Accelerating Neural Differential Equations by Regularizing Internal Solver Heuristics
May 09, 2021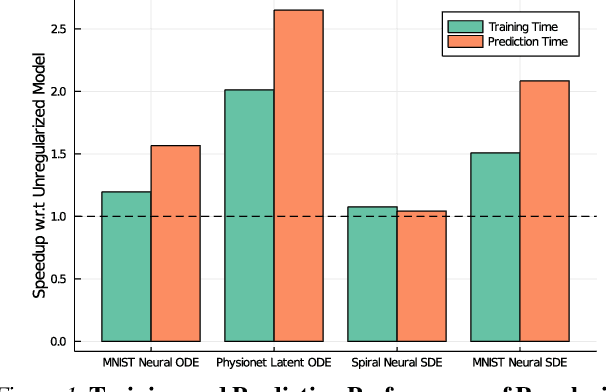
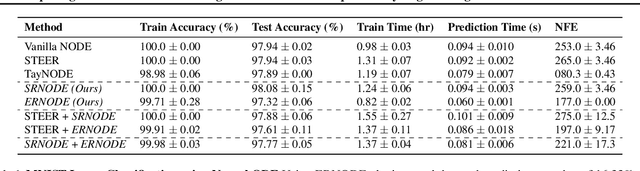
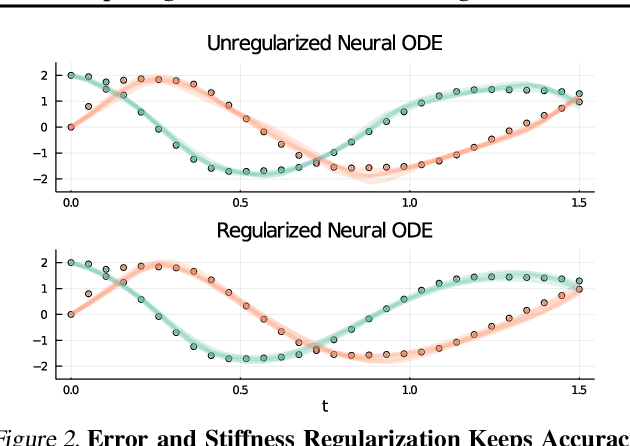
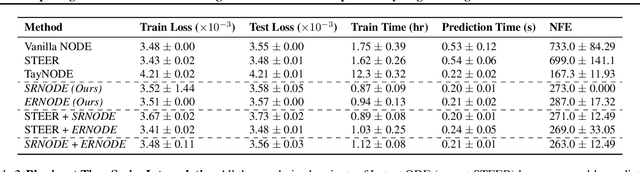
Abstract:Democratization of machine learning requires architectures that automatically adapt to new problems. Neural Differential Equations (NDEs) have emerged as a popular modeling framework by removing the need for ML practitioners to choose the number of layers in a recurrent model. While we can control the computational cost by choosing the number of layers in standard architectures, in NDEs the number of neural network evaluations for a forward pass can depend on the number of steps of the adaptive ODE solver. But, can we force the NDE to learn the version with the least steps while not increasing the training cost? Current strategies to overcome slow prediction require high order automatic differentiation, leading to significantly higher training time. We describe a novel regularization method that uses the internal cost heuristics of adaptive differential equation solvers combined with discrete adjoint sensitivities to guide the training process towards learning NDEs that are easier to solve. This approach opens up the blackbox numerical analysis behind the differential equation solver's algorithm and directly uses its local error estimates and stiffness heuristics as cheap and accurate cost estimates. We incorporate our method without any change in the underlying NDE framework and show that our method extends beyond Ordinary Differential Equations to accommodate Neural Stochastic Differential Equations. We demonstrate how our approach can halve the prediction time and, unlike other methods which can increase the training time by an order of magnitude, we demonstrate similar reduction in training times. Together this showcases how the knowledge embedded within state-of-the-art equation solvers can be used to enhance machine learning.
Accelerating Simulation of Stiff Nonlinear Systems using Continuous-Time Echo State Networks
Oct 19, 2020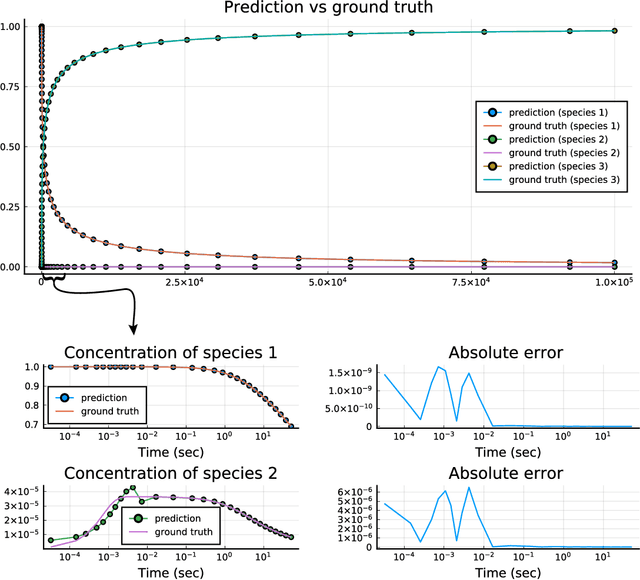
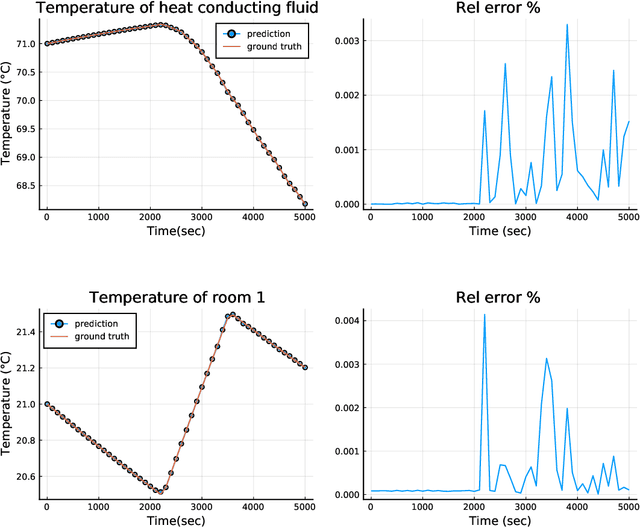
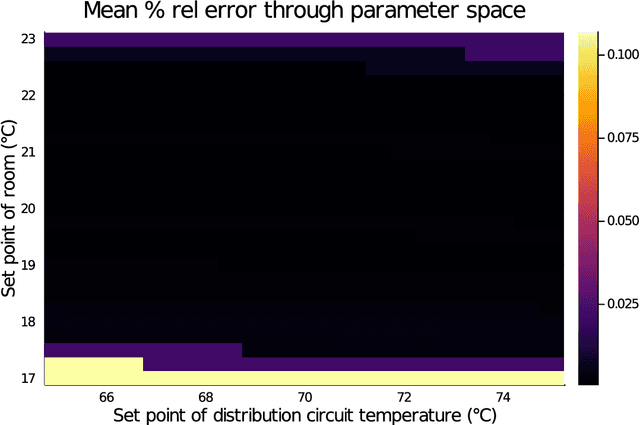
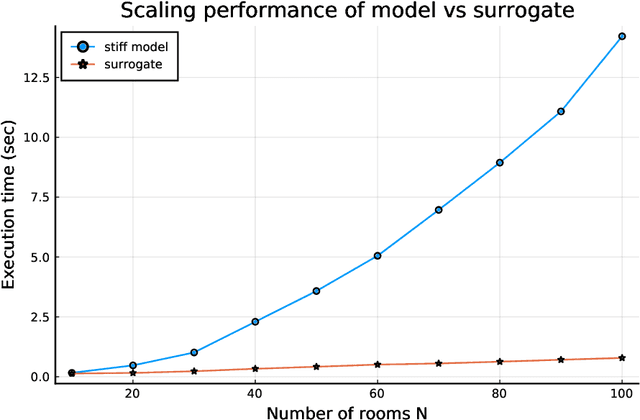
Abstract:Modern design, control, and optimization often requires simulation of highly nonlinear models, leading to prohibitive computational costs. These costs can be amortized by evaluating a cheap surrogate of the full model. Here we present a general data-driven method, the continuous-time echo state network (CTESN), for generating surrogates of nonlinear ordinary differential equations with dynamics at widely separated timescales. We empirically demonstrate near-constant time performance using our CTESNs on a physically motivated scalable model of a heating system whose full execution time increases exponentially, while maintaining relative error of within 0.2 %. We also show that our model captures fast transients as well as slow dynamics effectively, while other techniques such as physics informed neural networks have difficulties trying to train and predict the highly nonlinear behavior of these models.
Universal Differential Equations for Scientific Machine Learning
Jan 13, 2020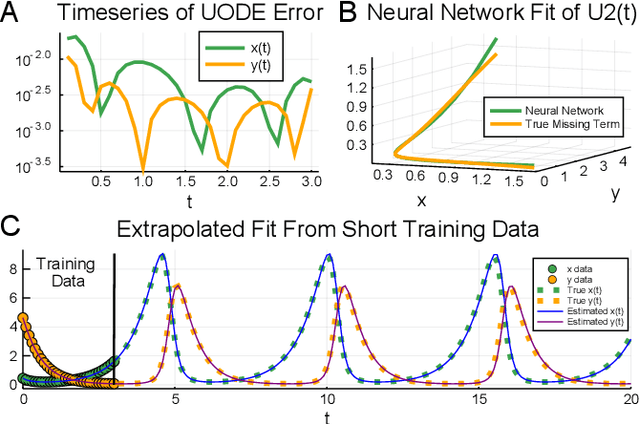
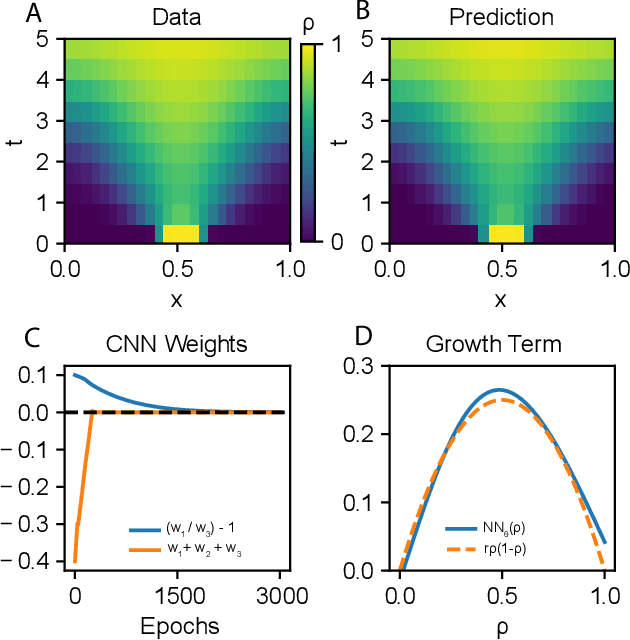
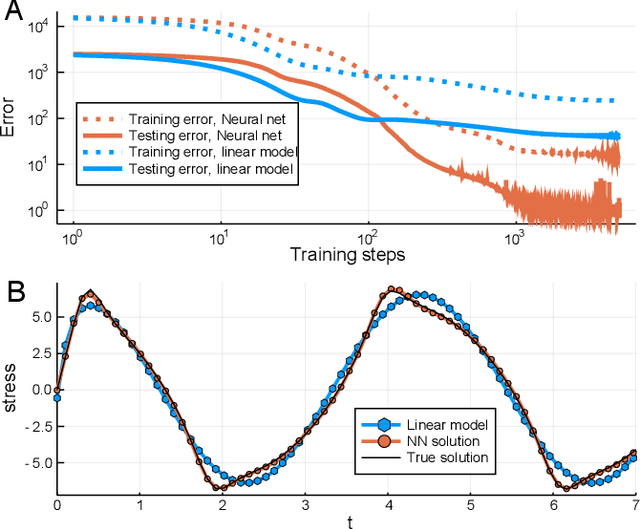
Abstract:In the context of science, the well-known adage "a picture is worth a thousand words" might well be "a model is worth a thousand datasets." Scientific models, such as Newtonian physics or biological gene regulatory networks, are human-driven simplifications of complex phenomena that serve as surrogates for the countless experiments that validated the models. Recently, machine learning has been able to overcome the inaccuracies of approximate modeling by directly learning the entire set of nonlinear interactions from data. However, without any predetermined structure from the scientific basis behind the problem, machine learning approaches are flexible but data-expensive, requiring large databases of homogeneous labeled training data. A central challenge is reconciling data that is at odds with simplified models without requiring "big data". In this work we develop a new methodology, universal differential equations (UDEs), which augments scientific models with machine-learnable structures for scientifically-based learning. We show how UDEs can be utilized to discover previously unknown governing equations, accurately extrapolate beyond the original data, and accelerate model simulation, all in a time and data-efficient manner. This advance is coupled with open-source software that allows for training UDEs which incorporate physical constraints, delayed interactions, implicitly-defined events, and intrinsic stochasticity in the model. Our examples show how a diverse set of computationally-difficult modeling issues across scientific disciplines, from automatically discovering biological mechanisms to accelerating climate simulations by 15,000x, can be handled by training UDEs.
DiffEqFlux.jl - A Julia Library for Neural Differential Equations
Feb 06, 2019Abstract:DiffEqFlux.jl is a library for fusing neural networks and differential equations. In this work we describe differential equations from the viewpoint of data science and discuss the complementary nature between machine learning models and differential equations. We demonstrate the ability to incorporate DifferentialEquations.jl-defined differential equation problems into a Flux-defined neural network, and vice versa. The advantages of being able to use the entire DifferentialEquations.jl suite for this purpose is demonstrated by counter examples where simple integration strategies fail, but the sophisticated integration strategies provided by the DifferentialEquations.jl library succeed. This is followed by a demonstration of delay differential equations and stochastic differential equations inside of neural networks. We show high-level functionality for defining neural ordinary differential equations (neural networks embedded into the differential equation) and describe the extra models in the Flux model zoo which includes neural stochastic differential equations. We conclude by discussing the various adjoint methods used for backpropogation of the differential equation solvers. DiffEqFlux.jl is an important contribution to the area, as it allows the full weight of the differential equation solvers developed from decades of research in the scientific computing field to be readily applied to the challenges posed by machine learning and data science.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge