Christopher Rackauckas
Semi-Explicit Neural DAEs: Learning Long-Horizon Dynamical Systems with Algebraic Constraints
May 26, 2025Abstract:Despite the promise of scientific machine learning (SciML) in combining data-driven techniques with mechanistic modeling, existing approaches for incorporating hard constraints in neural differential equations (NDEs) face significant limitations. Scalability issues and poor numerical properties prevent these neural models from being used for modeling physical systems with complicated conservation laws. We propose Manifold-Projected Neural ODEs (PNODEs), a method that explicitly enforces algebraic constraints by projecting each ODE step onto the constraint manifold. This framework arises naturally from semi-explicit differential-algebraic equations (DAEs), and includes both a robust iterative variant and a fast approximation requiring a single Jacobian factorization. We further demonstrate that prior works on relaxation methods are special cases of our approach. PNODEs consistently outperform baselines across six benchmark problems achieving a mean constraint violation error below $10^{-10}$. Additionally, PNODEs consistently achieve lower runtime compared to other methods for a given level of error tolerance. These results show that constraint projection offers a simple strategy for learning physically consistent long-horizon dynamics.
Differentiable Programming for Differential Equations: A Review
Jun 14, 2024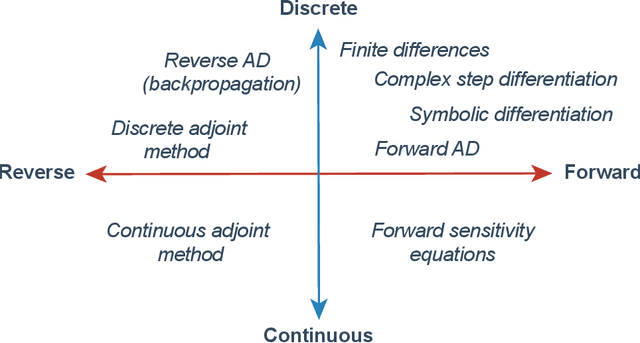
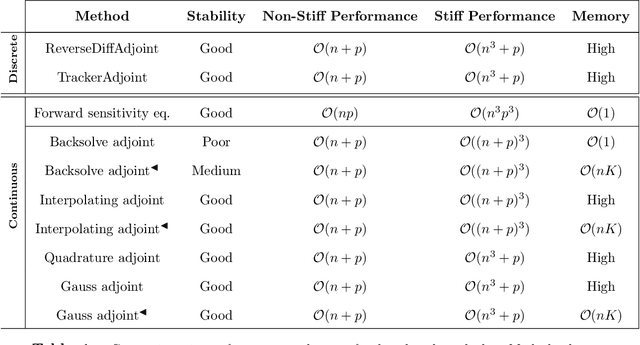
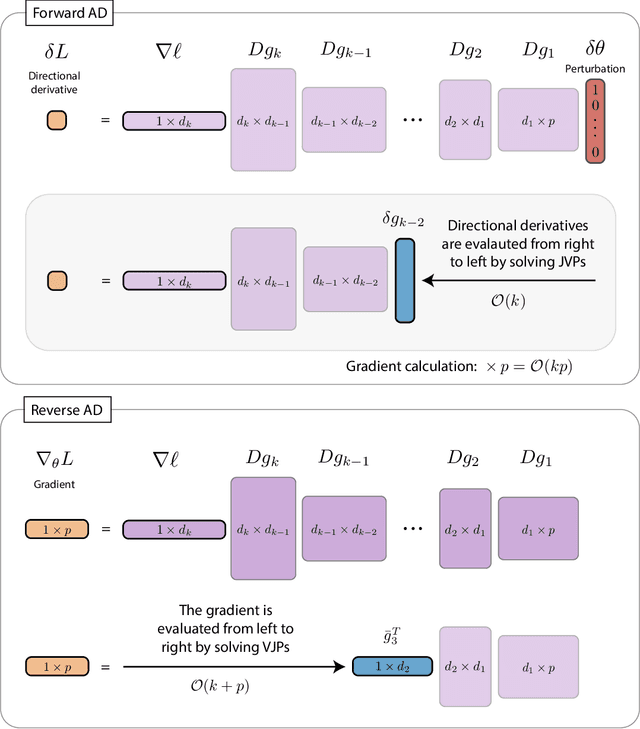
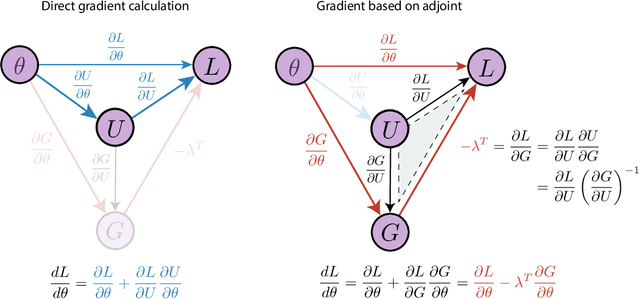
Abstract:The differentiable programming paradigm is a cornerstone of modern scientific computing. It refers to numerical methods for computing the gradient of a numerical model's output. Many scientific models are based on differential equations, where differentiable programming plays a crucial role in calculating model sensitivities, inverting model parameters, and training hybrid models that combine differential equations with data-driven approaches. Furthermore, recognizing the strong synergies between inverse methods and machine learning offers the opportunity to establish a coherent framework applicable to both fields. Differentiating functions based on the numerical solution of differential equations is non-trivial. Numerous methods based on a wide variety of paradigms have been proposed in the literature, each with pros and cons specific to the type of problem investigated. Here, we provide a comprehensive review of existing techniques to compute derivatives of numerical solutions of differential equations. We first discuss the importance of gradients of solutions of differential equations in a variety of scientific domains. Second, we lay out the mathematical foundations of the various approaches and compare them with each other. Third, we cover the computational considerations and explore the solutions available in modern scientific software. Last but not least, we provide best-practices and recommendations for practitioners. We hope that this work accelerates the fusion of scientific models and data, and fosters a modern approach to scientific modelling.
Differentiating Metropolis-Hastings to Optimize Intractable Densities
Jun 30, 2023Abstract:We develop an algorithm for automatic differentiation of Metropolis-Hastings samplers, allowing us to differentiate through probabilistic inference, even if the model has discrete components within it. Our approach fuses recent advances in stochastic automatic differentiation with traditional Markov chain coupling schemes, providing an unbiased and low-variance gradient estimator. This allows us to apply gradient-based optimization to objectives expressed as expectations over intractable target densities. We demonstrate our approach by finding an ambiguous observation in a Gaussian mixture model and by maximizing the specific heat in an Ising model.
Mixing Implicit and Explicit Deep Learning with Skip DEQs and Infinite Time Neural ODEs (Continuous DEQs)
Feb 04, 2022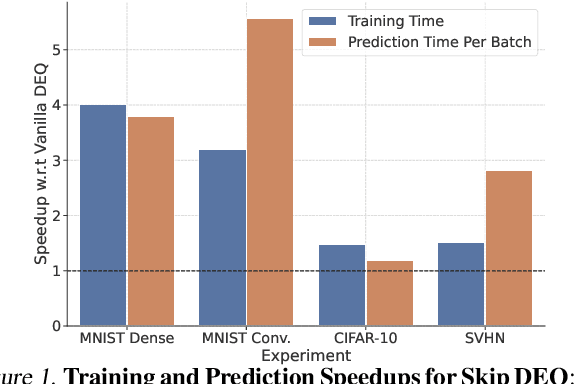
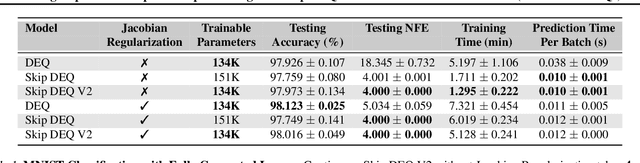
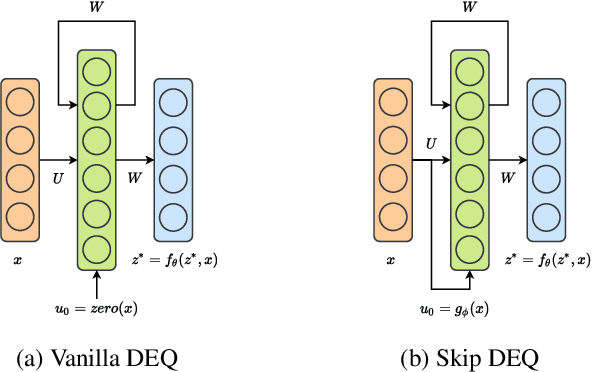
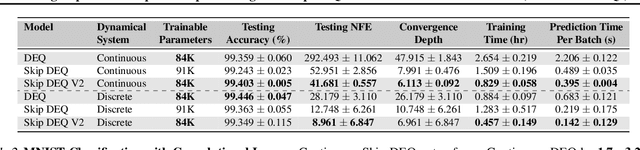
Abstract:Implicit deep learning architectures, like Neural ODEs and Deep Equilibrium Models (DEQs), separate the definition of a layer from the description of its solution process. While implicit layers allow features such as depth to adapt to new scenarios and inputs automatically, this adaptivity makes its computational expense challenging to predict. Numerous authors have noted that implicit layer techniques can be more computationally intensive than explicit layer methods. In this manuscript, we address the question: is there a way to simultaneously achieve the robustness of implicit layers while allowing the reduced computational expense of an explicit layer? To solve this we develop Skip DEQ, an implicit-explicit (IMEX) layer that simultaneously trains an explicit prediction followed by an implicit correction. We show that training this explicit layer is free and even decreases the training time by 2.5x and prediction time by 3.4x. We then further increase the "implicitness" of the DEQ by redefining the method in terms of an infinite time neural ODE which paradoxically decreases the training cost over a standard neural ODE by not requiring backpropagation through time. We demonstrate how the resulting Continuous Skip DEQ architecture trains more robustly than the original DEQ while achieving faster training and prediction times. Together, this manuscript shows how bridging the dichotomy of implicit and explicit deep learning can combine the advantages of both techniques.
High-performance symbolic-numerics via multiple dispatch
May 12, 2021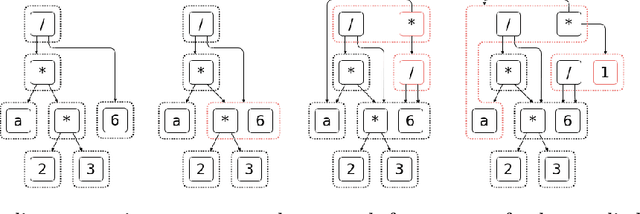
Abstract:As mathematical computing becomes more democratized in high-level languages, high-performance symbolic-numeric systems are necessary for domain scientists and engineers to get the best performance out of their machine without deep knowledge of code optimization. Naturally, users need different term types either to have different algebraic properties for them, or to use efficient data structures. To this end, we developed Symbolics.jl, an extendable symbolic system which uses dynamic multiple dispatch to change behavior depending on the domain needs. In this work we detail an underlying abstract term interface which allows for speed without sacrificing generality. We show that by formalizing a generic API on actions independent of implementation, we can retroactively add optimized data structures to our system without changing the pre-existing term rewriters. We showcase how this can be used to optimize term construction and give a 113x acceleration on general symbolic transformations. Further, we show that such a generic API allows for complementary term-rewriting implementations. We demonstrate the ability to swap between classical term-rewriting simplifiers and e-graph-based term-rewriting simplifiers. We showcase an e-graph ruleset which minimizes the number of CPU cycles during expression evaluation, and demonstrate how it simplifies a real-world reaction-network simulation to halve the runtime. Additionally, we show a reaction-diffusion partial differential equation solver which is able to be automatically converted into symbolic expressions via multiple dispatch tracing, which is subsequently accelerated and parallelized to give a 157x simulation speedup. Together, this presents Symbolics.jl as a next-generation symbolic-numeric computing environment geared towards modeling and simulation.
Opening the Blackbox: Accelerating Neural Differential Equations by Regularizing Internal Solver Heuristics
May 09, 2021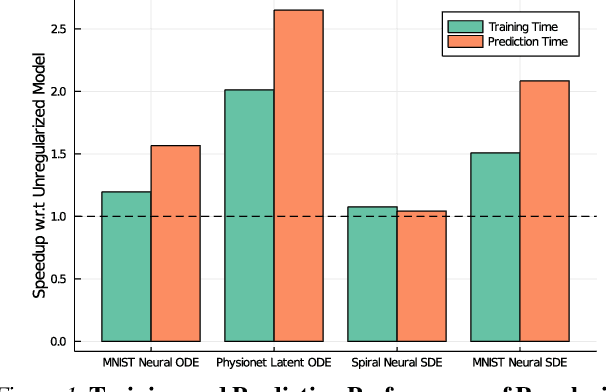
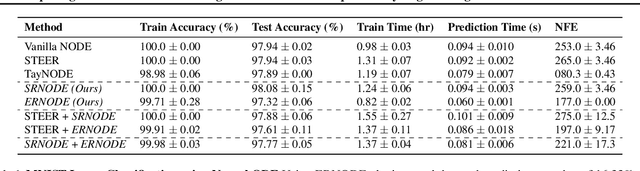
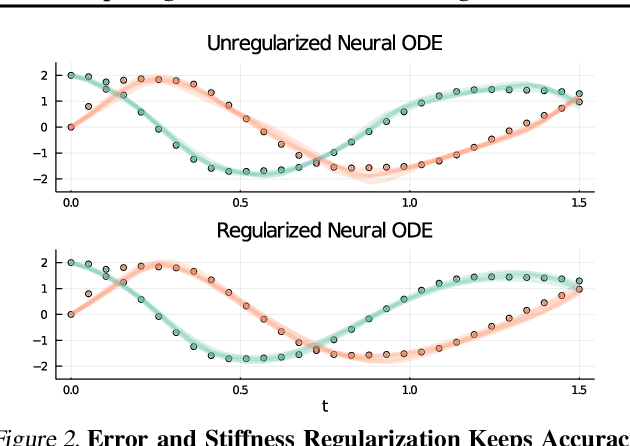
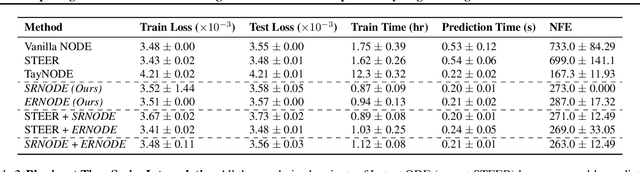
Abstract:Democratization of machine learning requires architectures that automatically adapt to new problems. Neural Differential Equations (NDEs) have emerged as a popular modeling framework by removing the need for ML practitioners to choose the number of layers in a recurrent model. While we can control the computational cost by choosing the number of layers in standard architectures, in NDEs the number of neural network evaluations for a forward pass can depend on the number of steps of the adaptive ODE solver. But, can we force the NDE to learn the version with the least steps while not increasing the training cost? Current strategies to overcome slow prediction require high order automatic differentiation, leading to significantly higher training time. We describe a novel regularization method that uses the internal cost heuristics of adaptive differential equation solvers combined with discrete adjoint sensitivities to guide the training process towards learning NDEs that are easier to solve. This approach opens up the blackbox numerical analysis behind the differential equation solver's algorithm and directly uses its local error estimates and stiffness heuristics as cheap and accurate cost estimates. We incorporate our method without any change in the underlying NDE framework and show that our method extends beyond Ordinary Differential Equations to accommodate Neural Stochastic Differential Equations. We demonstrate how our approach can halve the prediction time and, unlike other methods which can increase the training time by an order of magnitude, we demonstrate similar reduction in training times. Together this showcases how the knowledge embedded within state-of-the-art equation solvers can be used to enhance machine learning.
Stiff Neural Ordinary Differential Equations
Mar 29, 2021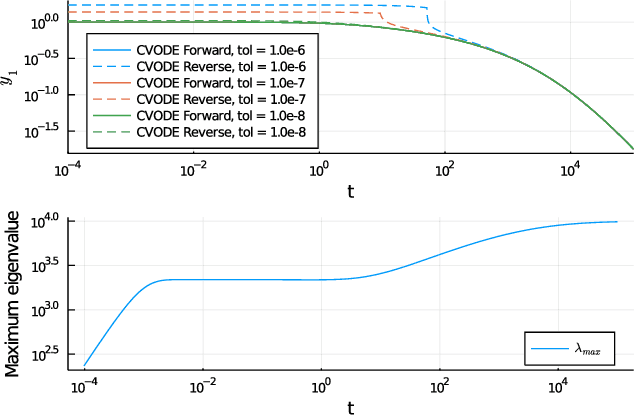
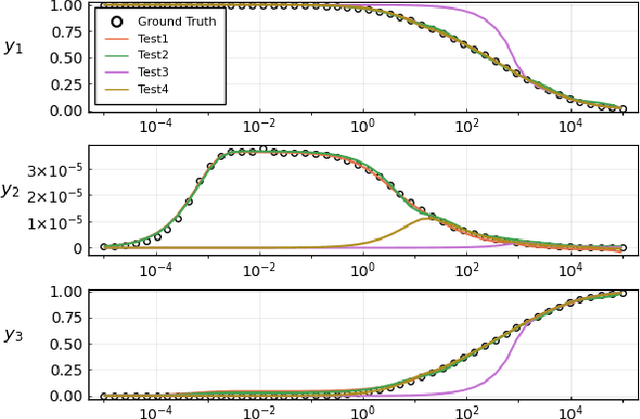
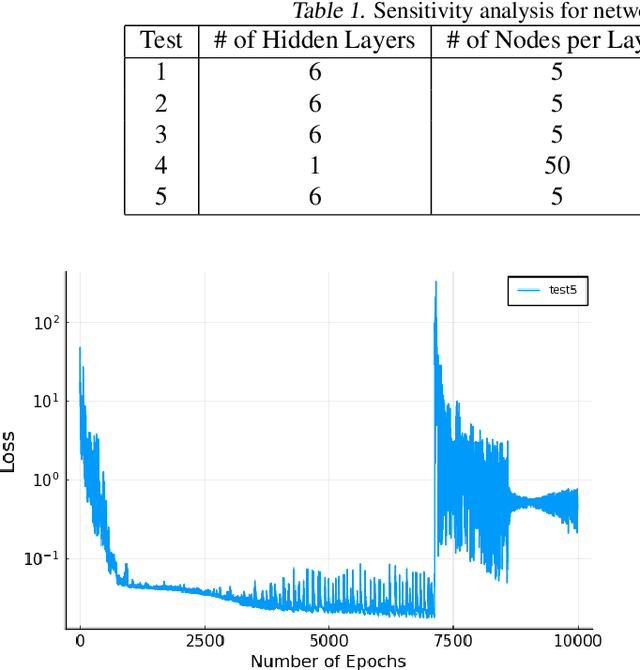
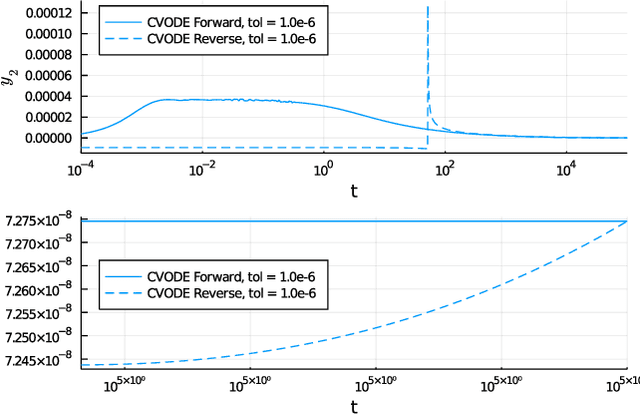
Abstract:Neural Ordinary Differential Equations (ODE) are a promising approach to learn dynamic models from time-series data in science and engineering applications. This work aims at learning Neural ODE for stiff systems, which are usually raised from chemical kinetic modeling in chemical and biological systems. We first show the challenges of learning neural ODE in the classical stiff ODE systems of Robertson's problem and propose techniques to mitigate the challenges associated with scale separations in stiff systems. We then present successful demonstrations in stiff systems of Robertson's problem and an air pollution problem. The demonstrations show that the usage of deep networks with rectified activations, proper scaling of the network outputs as well as loss functions, and stabilized gradient calculations are the key techniques enabling the learning of stiff neural ODE. The success of learning stiff neural ODE opens up possibilities of using neural ODEs in applications with widely varying time-scales, like chemical dynamics in energy conversion, environmental engineering, and the life sciences.
Signal Enhancement for Magnetic Navigation Challenge Problem
Jul 23, 2020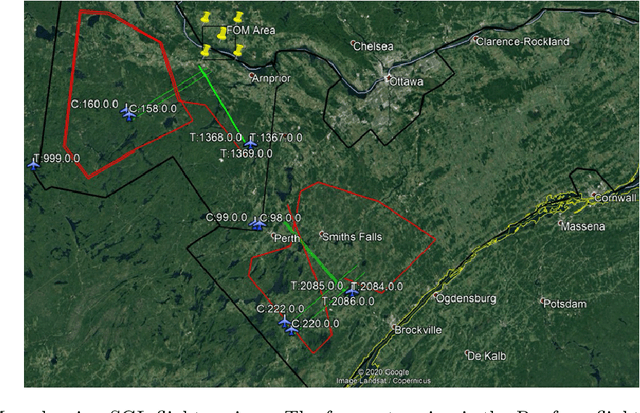
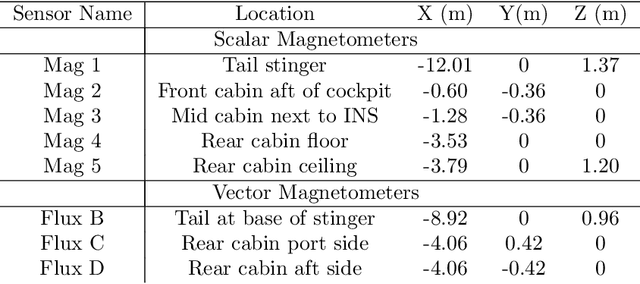
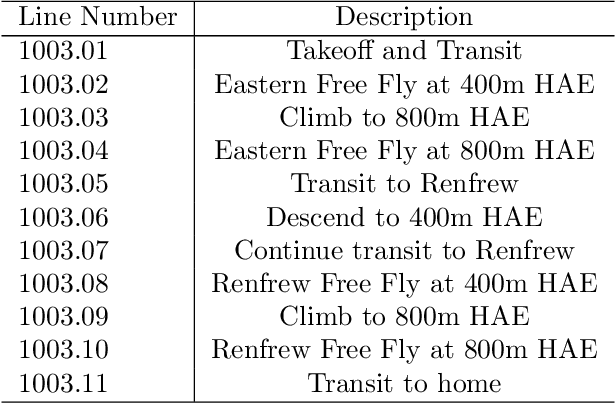
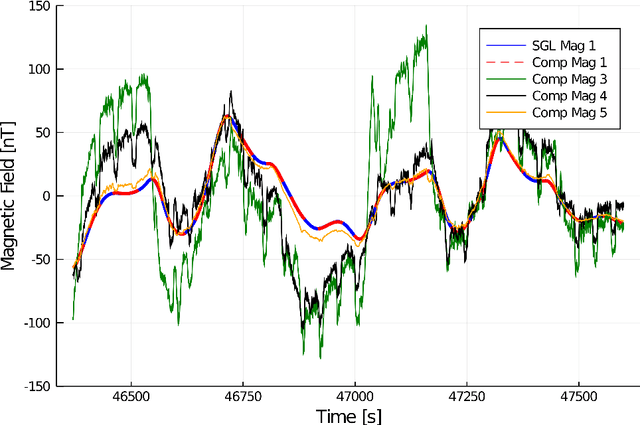
Abstract:Harnessing the magnetic field of the earth for navigation has shown promise as a viable alternative to other navigation systems. A magnetic navigation system collects its own magnetic field data using a magnetometer and uses magnetic anomaly maps to determine the current location. The greatest challenge with magnetic navigation arises when the magnetic field data from the magnetometer on the navigation system encompass the magnetic field from not just the earth, but also from the vehicle on which it is mounted. It is difficult to separate the earth magnetic anomaly field magnitude, which is crucial for navigation, from the total magnetic field magnitude reading from the sensor. The purpose of this challenge problem is to decouple the earth and aircraft magnetic signals in order to derive a clean signal from which to perform magnetic navigation. Baseline testing on the dataset shows that the earth magnetic field can be extracted from the total magnetic field using machine learning (ML). The challenge is to remove the aircraft magnetic field from the total magnetic field using a trained neural network. These challenges offer an opportunity to construct an effective neural network for removing the aircraft magnetic field from the dataset, using an ML algorithm integrated with physics of magnetic navigation.
Universal Differential Equations for Scientific Machine Learning
Jan 13, 2020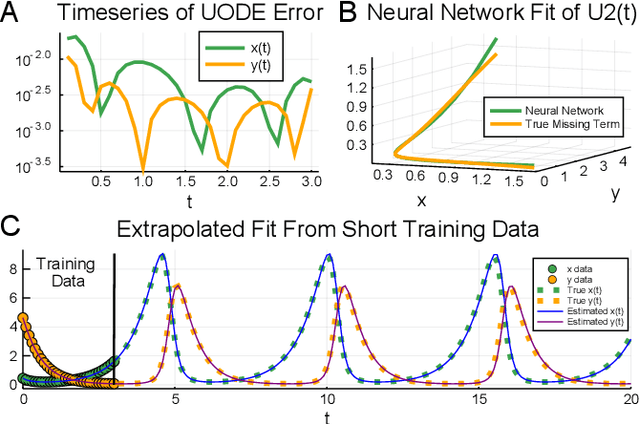
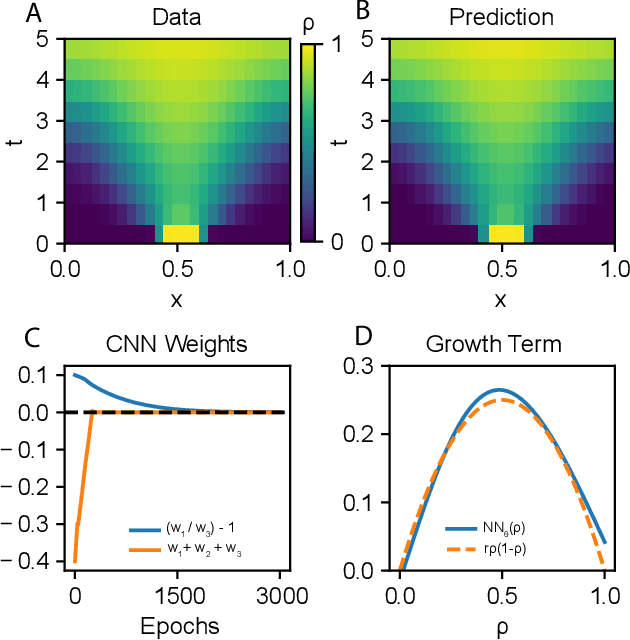
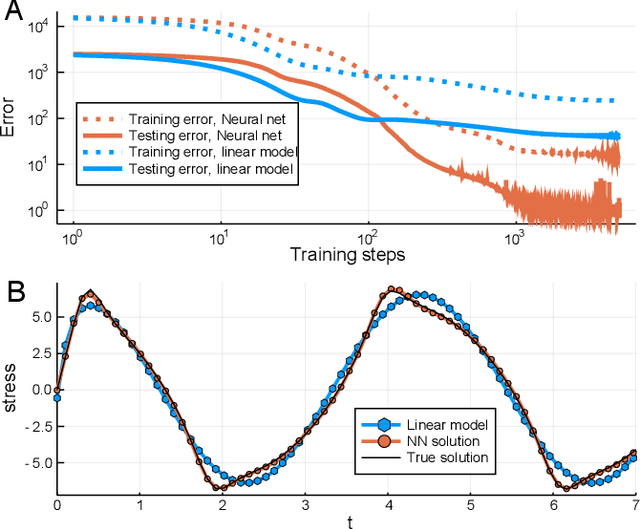
Abstract:In the context of science, the well-known adage "a picture is worth a thousand words" might well be "a model is worth a thousand datasets." Scientific models, such as Newtonian physics or biological gene regulatory networks, are human-driven simplifications of complex phenomena that serve as surrogates for the countless experiments that validated the models. Recently, machine learning has been able to overcome the inaccuracies of approximate modeling by directly learning the entire set of nonlinear interactions from data. However, without any predetermined structure from the scientific basis behind the problem, machine learning approaches are flexible but data-expensive, requiring large databases of homogeneous labeled training data. A central challenge is reconciling data that is at odds with simplified models without requiring "big data". In this work we develop a new methodology, universal differential equations (UDEs), which augments scientific models with machine-learnable structures for scientifically-based learning. We show how UDEs can be utilized to discover previously unknown governing equations, accurately extrapolate beyond the original data, and accelerate model simulation, all in a time and data-efficient manner. This advance is coupled with open-source software that allows for training UDEs which incorporate physical constraints, delayed interactions, implicitly-defined events, and intrinsic stochasticity in the model. Our examples show how a diverse set of computationally-difficult modeling issues across scientific disciplines, from automatically discovering biological mechanisms to accelerating climate simulations by 15,000x, can be handled by training UDEs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge