Victor Boussange
Deep learning-based species-area models reveal multi-scale patterns of species richness and turnover
Jul 08, 2025



Abstract:The number of species within ecosystems is influenced not only by their intrinsic characteristics but also by the spatial scale considered. As the sampled area expands, species richness increases, a phenomenon described by the species-area relationship (SAR). The accumulation dynamics of the SAR results from a complex interplay of biotic and abiotic processes operating at various spatial scales. However, the challenge of collecting exhaustive biodiversity records across spatial scales has hindered a comprehensive understanding of these dynamics. Here, we develop a deep learning approach that leverages sampling theory and small-scale ecological surveys to spatially resolve the scale-dependency of species richness. We demonstrate its performance by predicting the species richness of vascular plant communities across Europe, and evaluate the predictions against an independent dataset of plant community inventories. Our model improves species richness estimates by 32\% and delivers spatially explicit patterns of species richness and turnover for sampling areas ranging from square meters to hundreds of square kilometers. Explainable AI techniques further disentangle how drivers of species richness operate across spatial scales. The ability of our model to represent the multi-scale nature of biodiversity is essential to deliver robust biodiversity assessments and forecasts under global change.
Temporal horizons in forecasting: a performance-learnability trade-off
Jun 04, 2025



Abstract:When training autoregressive models for dynamical systems, a critical question arises: how far into the future should the model be trained to predict? Too short a horizon may miss long-term trends, while too long a horizon can impede convergence due to accumulating prediction errors. In this work, we formalize this trade-off by analyzing how the geometry of the loss landscape depends on the training horizon. We prove that for chaotic systems, the loss landscape's roughness grows exponentially with the training horizon, while for limit cycles, it grows linearly, making long-horizon training inherently challenging. However, we also show that models trained on long horizons generalize well to short-term forecasts, whereas those trained on short horizons suffer exponentially (resp. linearly) worse long-term predictions in chaotic (resp. periodic) systems. We validate our theory through numerical experiments and discuss practical implications for selecting training horizons. Our results provide a principled foundation for hyperparameter optimization in autoregressive forecasting models.
Differentiable Programming for Differential Equations: A Review
Jun 14, 2024
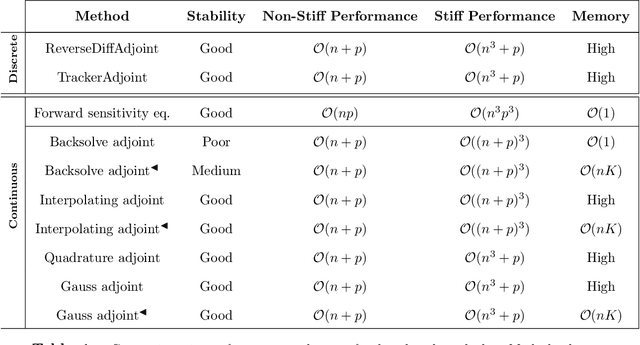
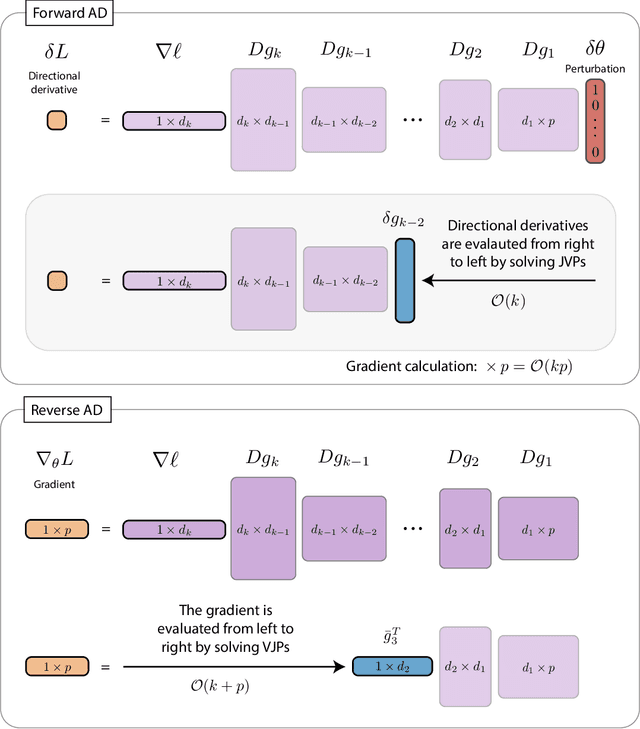
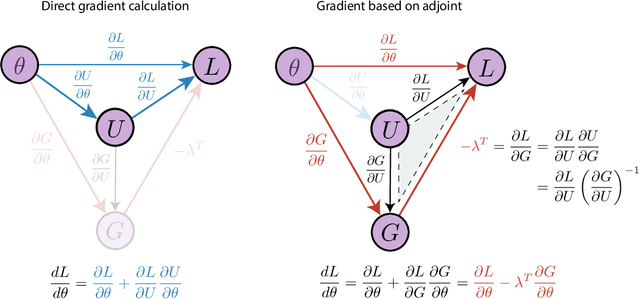
Abstract:The differentiable programming paradigm is a cornerstone of modern scientific computing. It refers to numerical methods for computing the gradient of a numerical model's output. Many scientific models are based on differential equations, where differentiable programming plays a crucial role in calculating model sensitivities, inverting model parameters, and training hybrid models that combine differential equations with data-driven approaches. Furthermore, recognizing the strong synergies between inverse methods and machine learning offers the opportunity to establish a coherent framework applicable to both fields. Differentiating functions based on the numerical solution of differential equations is non-trivial. Numerous methods based on a wide variety of paradigms have been proposed in the literature, each with pros and cons specific to the type of problem investigated. Here, we provide a comprehensive review of existing techniques to compute derivatives of numerical solutions of differential equations. We first discuss the importance of gradients of solutions of differential equations in a variety of scientific domains. Second, we lay out the mathematical foundations of the various approaches and compare them with each other. Third, we cover the computational considerations and explore the solutions available in modern scientific software. Last but not least, we provide best-practices and recommendations for practitioners. We hope that this work accelerates the fusion of scientific models and data, and fosters a modern approach to scientific modelling.
Deep learning approximations for non-local nonlinear PDEs with Neumann boundary conditions
May 07, 2022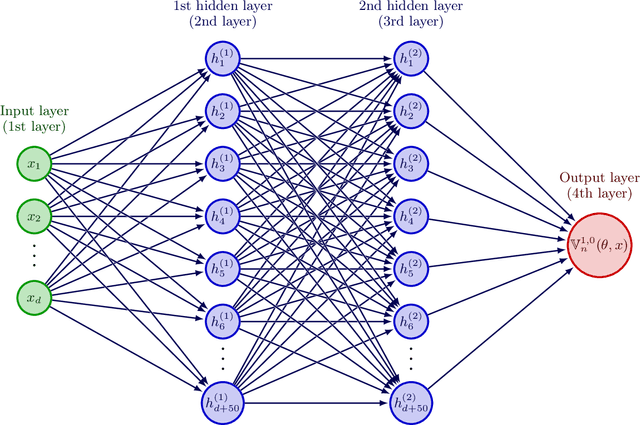
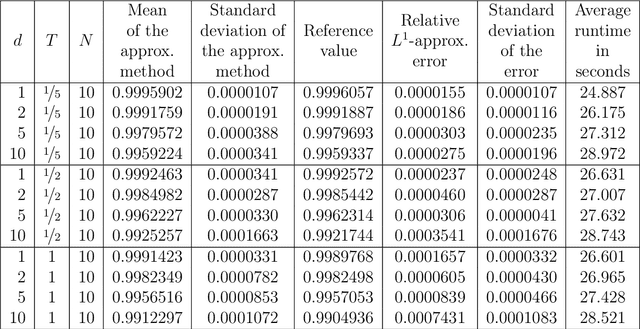
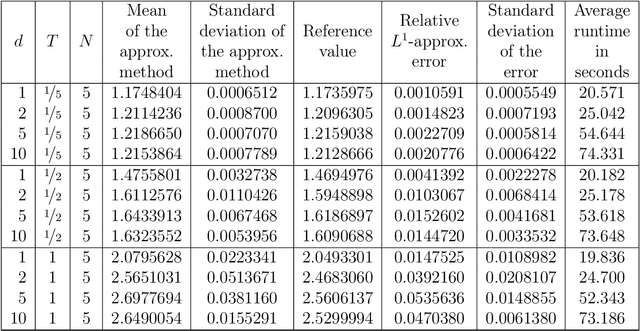
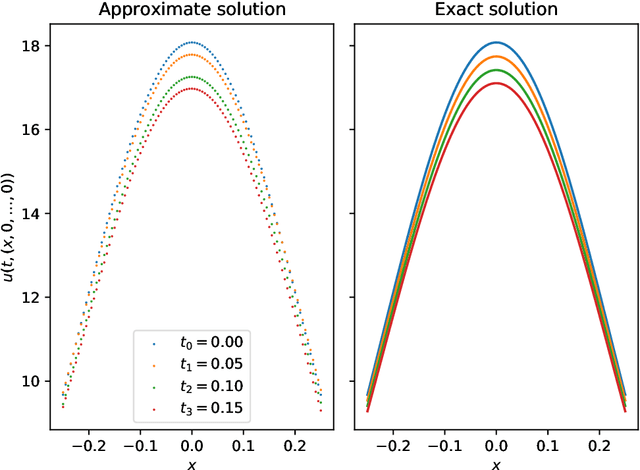
Abstract:Nonlinear partial differential equations (PDEs) are used to model dynamical processes in a large number of scientific fields, ranging from finance to biology. In many applications standard local models are not sufficient to accurately account for certain non-local phenomena such as, e.g., interactions at a distance. In order to properly capture these phenomena non-local nonlinear PDE models are frequently employed in the literature. In this article we propose two numerical methods based on machine learning and on Picard iterations, respectively, to approximately solve non-local nonlinear PDEs. The proposed machine learning-based method is an extended variant of a deep learning-based splitting-up type approximation method previously introduced in the literature and utilizes neural networks to provide approximate solutions on a subset of the spatial domain of the solution. The Picard iterations-based method is an extended variant of the so-called full history recursive multilevel Picard approximation scheme previously introduced in the literature and provides an approximate solution for a single point of the domain. Both methods are mesh-free and allow non-local nonlinear PDEs with Neumann boundary conditions to be solved in high dimensions. In the two methods, the numerical difficulties arising due to the dimensionality of the PDEs are avoided by (i) using the correspondence between the expected trajectory of reflected stochastic processes and the solution of PDEs (given by the Feynman-Kac formula) and by (ii) using a plain vanilla Monte Carlo integration to handle the non-local term. We evaluate the performance of the two methods on five different PDEs arising in physics and biology. In all cases, the methods yield good results in up to 10 dimensions with short run times. Our work extends recently developed methods to overcome the curse of dimensionality in solving PDEs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge