Yixin Wang
Causal Representation Meets Stochastic Modeling under Generic Geometry
Feb 04, 2026Abstract:Learning meaningful causal representations from observations has emerged as a crucial task for facilitating machine learning applications and driving scientific discoveries in fields such as climate science, biology, and physics. This process involves disentangling high-level latent variables and their causal relationships from low-level observations. Previous work in this area that achieves identifiability typically focuses on cases where the observations are either i.i.d. or follow a latent discrete-time process. Nevertheless, many real-world settings require identifying latent variables that are continuous-time stochastic processes (e.g., multivariate point processes). To this end, we develop identifiable causal representation learning for continuous-time latent stochastic point processes. We study its identifiability by analyzing the geometry of the parameter space. Furthermore, we develop MUTATE, an identifiable variational autoencoder framework with a time-adaptive transition module to infer stochastic dynamics. Across simulated and empirical studies, we find that MUTATE can effectively answer scientific questions, such as the accumulation of mutations in genomics and the mechanisms driving neuron spike triggers in response to time-varying dynamics.
Terminal-Bench: Benchmarking Agents on Hard, Realistic Tasks in Command Line Interfaces
Jan 17, 2026Abstract:AI agents may soon become capable of autonomously completing valuable, long-horizon tasks in diverse domains. Current benchmarks either do not measure real-world tasks, or are not sufficiently difficult to meaningfully measure frontier models. To this end, we present Terminal-Bench 2.0: a carefully curated hard benchmark composed of 89 tasks in computer terminal environments inspired by problems from real workflows. Each task features a unique environment, human-written solution, and comprehensive tests for verification. We show that frontier models and agents score less than 65\% on the benchmark and conduct an error analysis to identify areas for model and agent improvement. We publish the dataset and evaluation harness to assist developers and researchers in future work at https://www.tbench.ai/ .
Meta-probabilistic Modeling
Jan 08, 2026Abstract:While probabilistic graphical models can discover latent structure in data, their effectiveness hinges on choosing well-specified models. Identifying such models is challenging in practice, often requiring iterative checking and revision through trial and error. To this end, we propose meta-probabilistic modeling (MPM), a meta-learning algorithm that learns generative model structure directly from multiple related datasets. MPM uses a hierarchical architecture where global model specifications are shared across datasets while local parameters remain dataset-specific. For learning and inference, we propose a tractable VAE-inspired surrogate objective, and optimize it through bi-level optimization: local variables are updated analytically via coordinate ascent, while global parameters are trained with gradient-based methods. We evaluate MPM on object-centric image modeling and sequential text modeling, demonstrating that it adapts generative models to data while recovering meaningful latent representations.
On the Identifiability of Regime-Switching Models with Multi-Lag Dependencies
Jan 06, 2026Abstract:Identifiability is central to the interpretability of deep latent variable models, ensuring parameterisations are uniquely determined by the data-generating distribution. However, it remains underexplored for deep regime-switching time series. We develop a general theoretical framework for multi-lag Regime-Switching Models (RSMs), encompassing Markov Switching Models (MSMs) and Switching Dynamical Systems (SDSs). For MSMs, we formulate the model as a temporally structured finite mixture and prove identifiability of both the number of regimes and the multi-lag transitions in a nonlinear-Gaussian setting. For SDSs, we establish identifiability of the latent variables up to permutation and scaling via temporal structure, which in turn yields conditions for identifiability of regime-dependent latent causal graphs (up to regime/node permutations). Our results hold in a fully unsupervised setting through architectural and noise assumptions that are directly enforceable via neural network design. We complement the theory with a flexible variational estimator that satisfies the assumptions and validate the results on synthetic benchmarks. Across real-world datasets from neuroscience, finance, and climate, identifiability leads to more trustworthy interpretability analysis, which is crucial for scientific discovery.
Environment-Adaptive Covariate Selection: Learning When to Use Spurious Correlations for Out-of-Distribution Prediction
Jan 05, 2026Abstract:Out-of-distribution (OOD) prediction is often approached by restricting models to causal or invariant covariates, avoiding non-causal spurious associations that may be unstable across environments. Despite its theoretical appeal, this strategy frequently underperforms empirical risk minimization (ERM) in practice. We investigate the source of this gap and show that such failures naturally arise when only a subset of the true causes of the outcome is observed. In these settings, non-causal spurious covariates can serve as informative proxies for unobserved causes and substantially improve prediction, except under distribution shifts that break these proxy relationships. Consequently, the optimal set of predictive covariates is neither universal nor necessarily exhibits invariant relationships with the outcome across all environments, but instead depends on the specific type of shift encountered. Crucially, we observe that different covariate shifts induce distinct, observable signatures in the covariate distribution itself. Moreover, these signatures can be extracted from unlabeled data in the target OOD environment and used to assess when proxy covariates remain reliable and when they fail. Building on this observation, we propose an environment-adaptive covariate selection (EACS) algorithm that maps environment-level covariate summaries to environment-specific covariate sets, while allowing the incorporation of prior causal knowledge as constraints. Across simulations and applied datasets, EACS consistently outperforms static causal, invariant, and ERM-based predictors under diverse distribution shifts.
Last Layer Empirical Bayes
May 21, 2025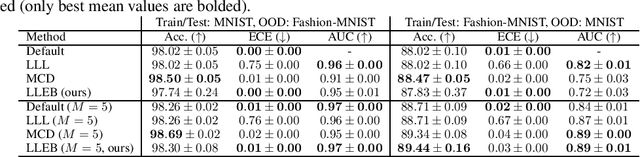
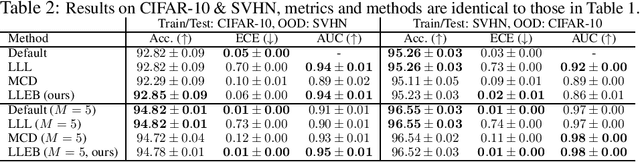

Abstract:The task of quantifying the inherent uncertainty associated with neural network predictions is a key challenge in artificial intelligence. Bayesian neural networks (BNNs) and deep ensembles are among the most prominent approaches to tackle this task. Both approaches produce predictions by computing an expectation of neural network outputs over some distribution on the corresponding weights; this distribution is given by the posterior in the case of BNNs, and by a mixture of point masses for ensembles. Inspired by recent work showing that the distribution used by ensembles can be understood as a posterior corresponding to a learned data-dependent prior, we propose last layer empirical Bayes (LLEB). LLEB instantiates a learnable prior as a normalizing flow, which is then trained to maximize the evidence lower bound; to retain tractability we use the flow only on the last layer. We show why LLEB is well motivated, and how it interpolates between standard BNNs and ensembles in terms of the strength of the prior that they use. LLEB performs on par with existing approaches, highlighting that empirical Bayes is a promising direction for future research in uncertainty quantification.
Let Me Grok for You: Accelerating Grokking via Embedding Transfer from a Weaker Model
Apr 17, 2025Abstract:''Grokking'' is a phenomenon where a neural network first memorizes training data and generalizes poorly, but then suddenly transitions to near-perfect generalization after prolonged training. While intriguing, this delayed generalization phenomenon compromises predictability and efficiency. Ideally, models should generalize directly without delay. To this end, this paper proposes GrokTransfer, a simple and principled method for accelerating grokking in training neural networks, based on the key observation that data embedding plays a crucial role in determining whether generalization is delayed. GrokTransfer first trains a smaller, weaker model to reach a nontrivial (but far from optimal) test performance. Then, the learned input embedding from this weaker model is extracted and used to initialize the embedding in the target, stronger model. We rigorously prove that, on a synthetic XOR task where delayed generalization always occurs in normal training, GrokTransfer enables the target model to generalize directly without delay. Moreover, we demonstrate that, across empirical studies of different tasks, GrokTransfer effectively reshapes the training dynamics and eliminates delayed generalization, for both fully-connected neural networks and Transformers.
Streamlining Biomedical Research with Specialized LLMs
Apr 15, 2025Abstract:In this paper, we propose a novel system that integrates state-of-the-art, domain-specific large language models with advanced information retrieval techniques to deliver comprehensive and context-aware responses. Our approach facilitates seamless interaction among diverse components, enabling cross-validation of outputs to produce accurate, high-quality responses enriched with relevant data, images, tables, and other modalities. We demonstrate the system's capability to enhance response precision by leveraging a robust question-answering model, significantly improving the quality of dialogue generation. The system provides an accessible platform for real-time, high-fidelity interactions, allowing users to benefit from efficient human-computer interaction, precise retrieval, and simultaneous access to a wide range of literature and data. This dramatically improves the research efficiency of professionals in the biomedical and pharmaceutical domains and facilitates faster, more informed decision-making throughout the R\&D process. Furthermore, the system proposed in this paper is available at https://synapse-chat.patsnap.com.
Deep Generative Models: Complexity, Dimensionality, and Approximation
Apr 01, 2025Abstract:Generative networks have shown remarkable success in learning complex data distributions, particularly in generating high-dimensional data from lower-dimensional inputs. While this capability is well-documented empirically, its theoretical underpinning remains unclear. One common theoretical explanation appeals to the widely accepted manifold hypothesis, which suggests that many real-world datasets, such as images and signals, often possess intrinsic low-dimensional geometric structures. Under this manifold hypothesis, it is widely believed that to approximate a distribution on a $d$-dimensional Riemannian manifold, the latent dimension needs to be at least $d$ or $d+1$. In this work, we show that this requirement on the latent dimension is not necessary by demonstrating that generative networks can approximate distributions on $d$-dimensional Riemannian manifolds from inputs of any arbitrary dimension, even lower than $d$, taking inspiration from the concept of space-filling curves. This approach, in turn, leads to a super-exponential complexity bound of the deep neural networks through expanded neurons. Our findings thus challenge the conventional belief on the relationship between input dimensionality and the ability of generative networks to model data distributions. This novel insight not only corroborates the practical effectiveness of generative networks in handling complex data structures, but also underscores a critical trade-off between approximation error, dimensionality, and model complexity.
Doubly robust identification of treatment effects from multiple environments
Mar 18, 2025Abstract:Practical and ethical constraints often require the use of observational data for causal inference, particularly in medicine and social sciences. Yet, observational datasets are prone to confounding, potentially compromising the validity of causal conclusions. While it is possible to correct for biases if the underlying causal graph is known, this is rarely a feasible ask in practical scenarios. A common strategy is to adjust for all available covariates, yet this approach can yield biased treatment effect estimates, especially when post-treatment or unobserved variables are present. We propose RAMEN, an algorithm that produces unbiased treatment effect estimates by leveraging the heterogeneity of multiple data sources without the need to know or learn the underlying causal graph. Notably, RAMEN achieves doubly robust identification: it can identify the treatment effect whenever the causal parents of the treatment or those of the outcome are observed, and the node whose parents are observed satisfies an invariance assumption. Empirical evaluations on synthetic and real-world datasets show that our approach outperforms existing methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge