Min Woo Sun
Do VLMs Perceive or Recall? Probing Visual Perception vs. Memory with Classic Visual Illusions
Jan 29, 2026Abstract:Large Vision-Language Models (VLMs) often answer classic visual illusions "correctly" on original images, yet persist with the same responses when illusion factors are inverted, even though the visual change is obvious to humans. This raises a fundamental question: do VLMs perceive visual changes or merely recall memorized patterns? While several studies have noted this phenomenon, the underlying causes remain unclear. To move from observations to systematic understanding, this paper introduces VI-Probe, a controllable visual-illusion framework with graded perturbations and matched visual controls (without illusion inducer) that disentangles visually grounded perception from language-driven recall. Unlike prior work that focuses on averaged accuracy, we measure stability and sensitivity using Polarity-Flip Consistency, Template Fixation Index, and an illusion multiplier normalized against matched controls. Experiments across different families reveal that response persistence arises from heterogeneous causes rather than a single mechanism. For instance, GPT-5 exhibits memory override, Claude-Opus-4.1 shows perception-memory competition, while Qwen variants suggest visual-processing limits. Our findings challenge single-cause views and motivate probing-based evaluation that measures both knowledge and sensitivity to controlled visual change. Data and code are available at https://sites.google.com/view/vi-probe/.
PaperSearchQA: Learning to Search and Reason over Scientific Papers with RLVR
Jan 26, 2026Abstract:Search agents are language models (LMs) that reason and search knowledge bases (or the web) to answer questions; recent methods supervise only the final answer accuracy using reinforcement learning with verifiable rewards (RLVR). Most RLVR search agents tackle general-domain QA, which limits their relevance to technical AI systems in science, engineering, and medicine. In this work we propose training agents to search and reason over scientific papers -- this tests technical question-answering, it is directly relevant to real scientists, and the capabilities will be crucial to future AI Scientist systems. Concretely, we release a search corpus of 16 million biomedical paper abstracts and construct a challenging factoid QA dataset called PaperSearchQA with 60k samples answerable from the corpus, along with benchmarks. We train search agents in this environment to outperform non-RL retrieval baselines; we also perform further quantitative analysis and observe interesting agent behaviors like planning, reasoning, and self-verification. Our corpus, datasets, and benchmarks are usable with the popular Search-R1 codebase for RLVR training and released on https://huggingface.co/collections/jmhb/papersearchqa. Finally, our data creation methods are scalable and easily extendable to other scientific domains.
Can Large Language Models Match the Conclusions of Systematic Reviews?
May 28, 2025Abstract:Systematic reviews (SR), in which experts summarize and analyze evidence across individual studies to provide insights on a specialized topic, are a cornerstone for evidence-based clinical decision-making, research, and policy. Given the exponential growth of scientific articles, there is growing interest in using large language models (LLMs) to automate SR generation. However, the ability of LLMs to critically assess evidence and reason across multiple documents to provide recommendations at the same proficiency as domain experts remains poorly characterized. We therefore ask: Can LLMs match the conclusions of systematic reviews written by clinical experts when given access to the same studies? To explore this question, we present MedEvidence, a benchmark pairing findings from 100 SRs with the studies they are based on. We benchmark 24 LLMs on MedEvidence, including reasoning, non-reasoning, medical specialist, and models across varying sizes (from 7B-700B). Through our systematic evaluation, we find that reasoning does not necessarily improve performance, larger models do not consistently yield greater gains, and knowledge-based fine-tuning degrades accuracy on MedEvidence. Instead, most models exhibit similar behavior: performance tends to degrade as token length increases, their responses show overconfidence, and, contrary to human experts, all models show a lack of scientific skepticism toward low-quality findings. These results suggest that more work is still required before LLMs can reliably match the observations from expert-conducted SRs, even though these systems are already deployed and being used by clinicians. We release our codebase and benchmark to the broader research community to further investigate LLM-based SR systems.
MedCaseReasoning: Evaluating and learning diagnostic reasoning from clinical case reports
May 16, 2025Abstract:Doctors and patients alike increasingly use Large Language Models (LLMs) to diagnose clinical cases. However, unlike domains such as math or coding, where correctness can be objectively defined by the final answer, medical diagnosis requires both the outcome and the reasoning process to be accurate. Currently, widely used medical benchmarks like MedQA and MMLU assess only accuracy in the final answer, overlooking the quality and faithfulness of the clinical reasoning process. To address this limitation, we introduce MedCaseReasoning, the first open-access dataset for evaluating LLMs on their ability to align with clinician-authored diagnostic reasoning. The dataset includes 14,489 diagnostic question-and-answer cases, each paired with detailed reasoning statements derived from open-access medical case reports. We evaluate state-of-the-art reasoning LLMs on MedCaseReasoning and find significant shortcomings in their diagnoses and reasoning: for instance, the top-performing open-source model, DeepSeek-R1, achieves only 48% 10-shot diagnostic accuracy and mentions only 64% of the clinician reasoning statements (recall). However, we demonstrate that fine-tuning LLMs on the reasoning traces derived from MedCaseReasoning significantly improves diagnostic accuracy and clinical reasoning recall by an average relative gain of 29% and 41%, respectively. The open-source dataset, code, and models are available at https://github.com/kevinwu23/Stanford-MedCaseReasoning.
BIOMEDICA: An Open Biomedical Image-Caption Archive, Dataset, and Vision-Language Models Derived from Scientific Literature
Jan 14, 2025Abstract:The development of vision-language models (VLMs) is driven by large-scale and diverse multimodal datasets. However, progress toward generalist biomedical VLMs is limited by the lack of annotated, publicly accessible datasets across biology and medicine. Existing efforts are restricted to narrow domains, missing the full diversity of biomedical knowledge encoded in scientific literature. To address this gap, we introduce BIOMEDICA, a scalable, open-source framework to extract, annotate, and serialize the entirety of the PubMed Central Open Access subset into an easy-to-use, publicly accessible dataset. Our framework produces a comprehensive archive with over 24 million unique image-text pairs from over 6 million articles. Metadata and expert-guided annotations are also provided. We demonstrate the utility and accessibility of our resource by releasing BMCA-CLIP, a suite of CLIP-style models continuously pre-trained on the BIOMEDICA dataset via streaming, eliminating the need to download 27 TB of data locally. On average, our models achieve state-of-the-art performance across 40 tasks - spanning pathology, radiology, ophthalmology, dermatology, surgery, molecular biology, parasitology, and cell biology - excelling in zero-shot classification with a 6.56% average improvement (as high as 29.8% and 17.5% in dermatology and ophthalmology, respectively), and stronger image-text retrieval, all while using 10x less compute. To foster reproducibility and collaboration, we release our codebase and dataset for the broader research community.
Confidence intervals for the Cox model test error from cross-validation
Jan 26, 2022Abstract:Cross-validation (CV) is one of the most widely used techniques in statistical learning for estimating the test error of a model, but its behavior is not yet fully understood. It has been shown that standard confidence intervals for test error using estimates from CV may have coverage below nominal levels. This phenomenon occurs because each sample is used in both the training and testing procedures during CV and as a result, the CV estimates of the errors become correlated. Without accounting for this correlation, the estimate of the variance is smaller than it should be. One way to mitigate this issue is by estimating the mean squared error of the prediction error instead using nested CV. This approach has been shown to achieve superior coverage compared to intervals derived from standard CV. In this work, we generalize the nested CV idea to the Cox proportional hazards model and explore various choices of test error for this setting.
Training an Emotion Detection Classifier using Frames from a Mobile Therapeutic Game for Children with Developmental Disorders
Dec 16, 2020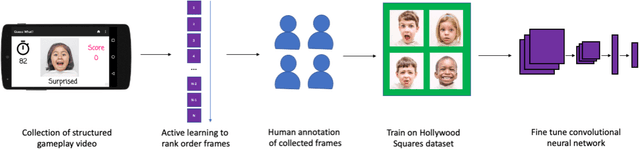
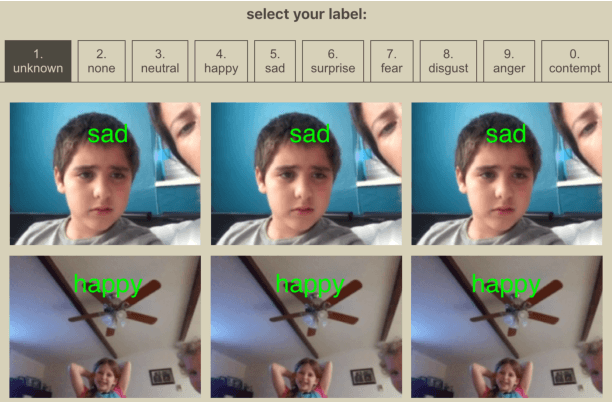
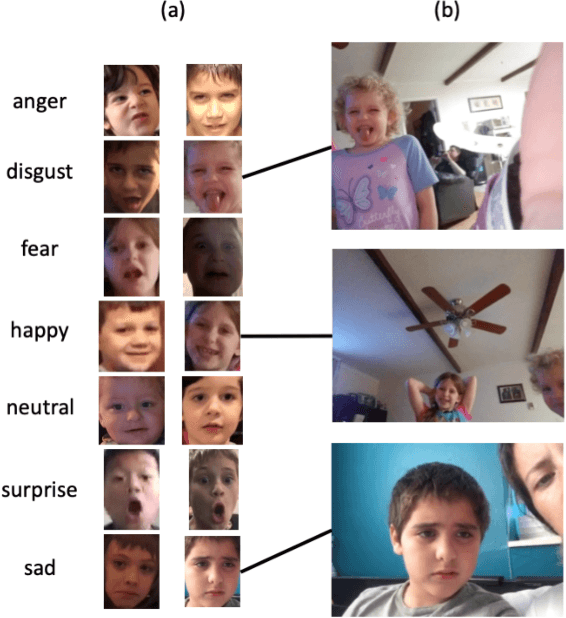
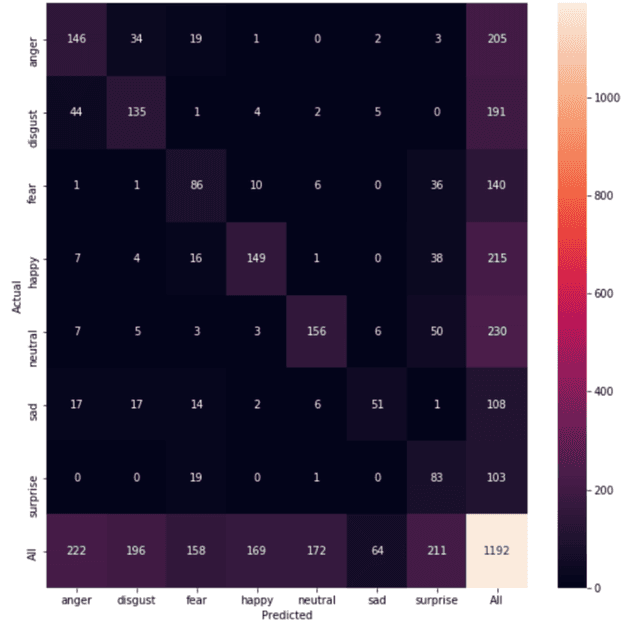
Abstract:Automated emotion classification could aid those who struggle to recognize emotion, including children with developmental behavioral conditions such as autism. However, most computer vision emotion models are trained on adult affect and therefore underperform on child faces. In this study, we designed a strategy to gamify the collection and the labeling of child affect data in an effort to boost the performance of automatic child emotion detection to a level closer to what will be needed for translational digital healthcare. We leveraged our therapeutic smartphone game, GuessWhat, which was designed in large part for children with developmental and behavioral conditions, to gamify the secure collection of video data of children expressing a variety of emotions prompted by the game. Through a secure web interface gamifying the human labeling effort, we gathered and labeled 2,155 videos, 39,968 emotion frames, and 106,001 labels on all images. With this drastically expanded pediatric emotion centric database (>30x larger than existing public pediatric affect datasets), we trained a pediatric emotion classification convolutional neural network (CNN) classifier of happy, sad, surprised, fearful, angry, disgust, and neutral expressions in children. The classifier achieved 66.9% balanced accuracy and 67.4% F1-score on the entirety of CAFE as well as 79.1% balanced accuracy and 78.0% F1-score on CAFE Subset A, a subset containing at least 60% human agreement on emotions labels. This performance is at least 10% higher than all previously published classifiers, the best of which reached 56.% balanced accuracy even when combining "anger" and "disgust" into a single class. This work validates that mobile games designed for pediatric therapies can generate high volumes of domain-relevant datasets to train state of the art classifiers to perform tasks highly relevant to precision health efforts.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge