Zepeng Huo
A Smart-Glasses for Emergency Medical Services via Multimodal Multitask Learning
Nov 17, 2025Abstract:Emergency Medical Technicians (EMTs) operate in high-pressure environments, making rapid, life-critical decisions under heavy cognitive and operational loads. We present EMSGlass, a smart-glasses system powered by EMSNet, the first multimodal multitask model for Emergency Medical Services (EMS), and EMSServe, a low-latency multimodal serving framework tailored to EMS scenarios. EMSNet integrates text, vital signs, and scene images to construct a unified real-time understanding of EMS incidents. Trained on real-world multimodal EMS datasets, EMSNet simultaneously supports up to five critical EMS tasks with superior accuracy compared to state-of-the-art unimodal baselines. Built on top of PyTorch, EMSServe introduces a modality-aware model splitter and a feature caching mechanism, achieving adaptive and efficient inference across heterogeneous hardware while addressing the challenge of asynchronous modality arrival in the field. By optimizing multimodal inference execution in EMS scenarios, EMSServe achieves 1.9x -- 11.7x speedup over direct PyTorch multimodal inference. A user study evaluation with six professional EMTs demonstrates that EMSGlass enhances real-time situational awareness, decision-making speed, and operational efficiency through intuitive on-glass interaction. In addition, qualitative insights from the user study provide actionable directions for extending EMSGlass toward next-generation AI-enabled EMS systems, bridging multimodal intelligence with real-world emergency response workflows.
Time-to-Event Pretraining for 3D Medical Imaging
Nov 14, 2024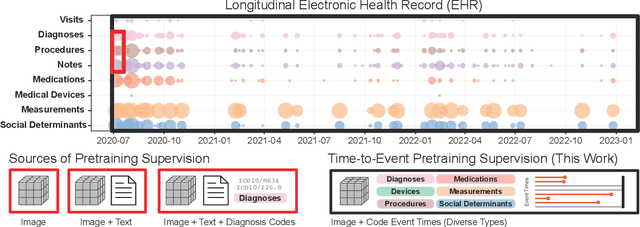
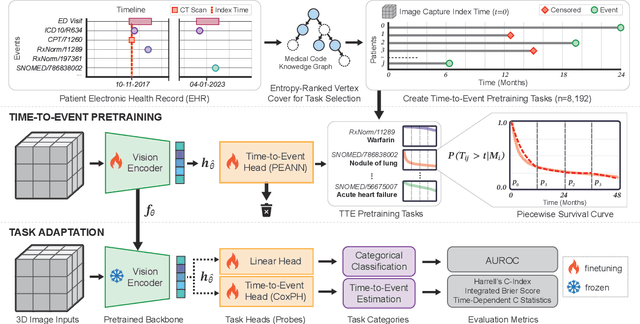
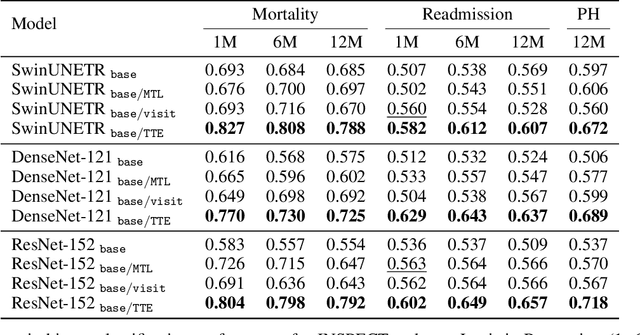
Abstract:With the rise of medical foundation models and the growing availability of imaging data, scalable pretraining techniques offer a promising way to identify imaging biomarkers predictive of future disease risk. While current self-supervised methods for 3D medical imaging models capture local structural features like organ morphology, they fail to link pixel biomarkers with long-term health outcomes due to a missing context problem. Current approaches lack the temporal context necessary to identify biomarkers correlated with disease progression, as they rely on supervision derived only from images and concurrent text descriptions. To address this, we introduce time-to-event pretraining, a pretraining framework for 3D medical imaging models that leverages large-scale temporal supervision from paired, longitudinal electronic health records (EHRs). Using a dataset of 18,945 CT scans (4.2 million 2D images) and time-to-event distributions across thousands of EHR-derived tasks, our method improves outcome prediction, achieving an average AUROC increase of 23.7% and a 29.4% gain in Harrell's C-index across 8 benchmark tasks. Importantly, these gains are achieved without sacrificing diagnostic classification performance. This study lays the foundation for integrating longitudinal EHR and 3D imaging data to advance clinical risk prediction.
Merlin: A Vision Language Foundation Model for 3D Computed Tomography
Jun 10, 2024



Abstract:Over 85 million computed tomography (CT) scans are performed annually in the US, of which approximately one quarter focus on the abdomen. Given the current radiologist shortage, there is a large impetus to use artificial intelligence to alleviate the burden of interpreting these complex imaging studies. Prior state-of-the-art approaches for automated medical image interpretation leverage vision language models (VLMs). However, current medical VLMs are generally limited to 2D images and short reports, and do not leverage electronic health record (EHR) data for supervision. We introduce Merlin - a 3D VLM that we train using paired CT scans (6+ million images from 15,331 CTs), EHR diagnosis codes (1.8+ million codes), and radiology reports (6+ million tokens). We evaluate Merlin on 6 task types and 752 individual tasks. The non-adapted (off-the-shelf) tasks include zero-shot findings classification (31 findings), phenotype classification (692 phenotypes), and zero-shot cross-modal retrieval (image to findings and image to impressions), while model adapted tasks include 5-year disease prediction (6 diseases), radiology report generation, and 3D semantic segmentation (20 organs). We perform internal validation on a test set of 5,137 CTs, and external validation on 7,000 clinical CTs and on two public CT datasets (VerSe, TotalSegmentator). Beyond these clinically-relevant evaluations, we assess the efficacy of various network architectures and training strategies to depict that Merlin has favorable performance to existing task-specific baselines. We derive data scaling laws to empirically assess training data needs for requisite downstream task performance. Furthermore, unlike conventional VLMs that require hundreds of GPUs for training, we perform all training on a single GPU.
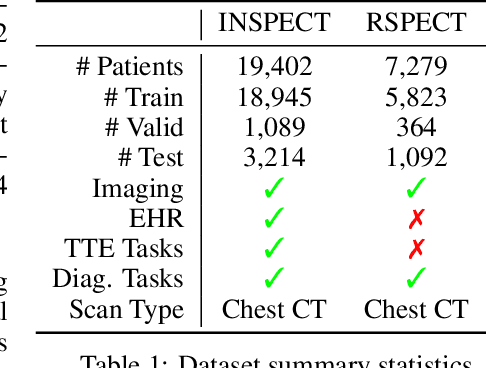
INSPECT: A Multimodal Dataset for Pulmonary Embolism Diagnosis and Prognosis
Nov 17, 2023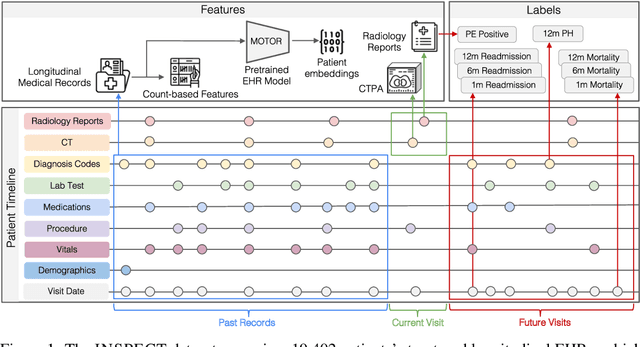
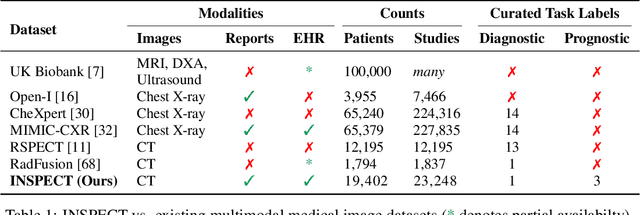
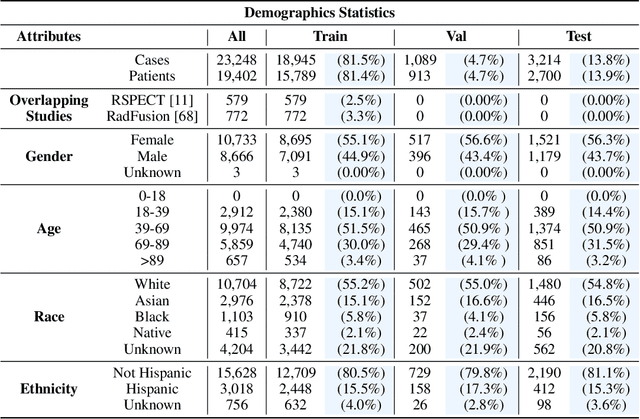
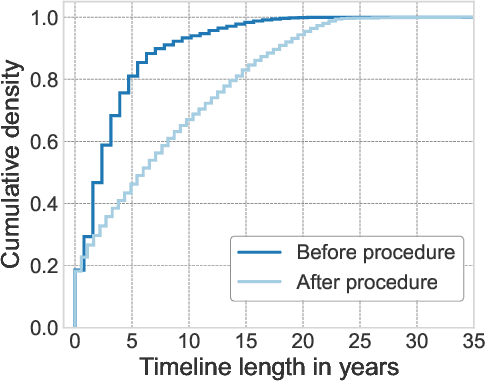
Abstract:Synthesizing information from multiple data sources plays a crucial role in the practice of modern medicine. Current applications of artificial intelligence in medicine often focus on single-modality data due to a lack of publicly available, multimodal medical datasets. To address this limitation, we introduce INSPECT, which contains de-identified longitudinal records from a large cohort of patients at risk for pulmonary embolism (PE), along with ground truth labels for multiple outcomes. INSPECT contains data from 19,402 patients, including CT images, radiology report impression sections, and structured electronic health record (EHR) data (i.e. demographics, diagnoses, procedures, vitals, and medications). Using INSPECT, we develop and release a benchmark for evaluating several baseline modeling approaches on a variety of important PE related tasks. We evaluate image-only, EHR-only, and multimodal fusion models. Trained models and the de-identified dataset are made available for non-commercial use under a data use agreement. To the best of our knowledge, INSPECT is the largest multimodal dataset integrating 3D medical imaging and EHR for reproducible methods evaluation and research.
MedAlign: A Clinician-Generated Dataset for Instruction Following with Electronic Medical Records
Aug 27, 2023
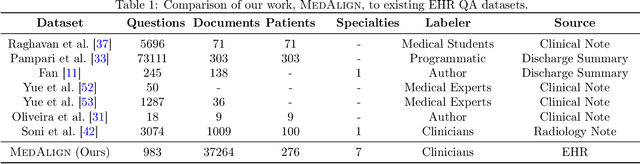
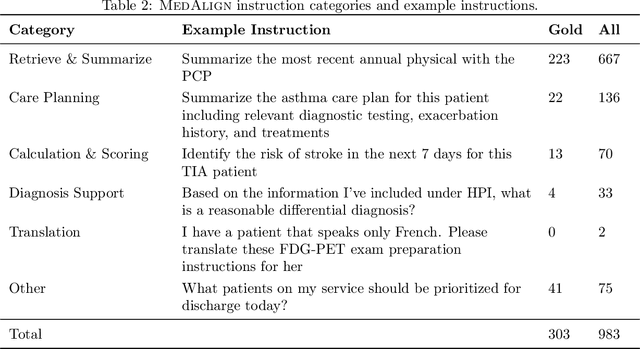

Abstract:The ability of large language models (LLMs) to follow natural language instructions with human-level fluency suggests many opportunities in healthcare to reduce administrative burden and improve quality of care. However, evaluating LLMs on realistic text generation tasks for healthcare remains challenging. Existing question answering datasets for electronic health record (EHR) data fail to capture the complexity of information needs and documentation burdens experienced by clinicians. To address these challenges, we introduce MedAlign, a benchmark dataset of 983 natural language instructions for EHR data. MedAlign is curated by 15 clinicians (7 specialities), includes clinician-written reference responses for 303 instructions, and provides 276 longitudinal EHRs for grounding instruction-response pairs. We used MedAlign to evaluate 6 general domain LLMs, having clinicians rank the accuracy and quality of each LLM response. We found high error rates, ranging from 35% (GPT-4) to 68% (MPT-7B-Instruct), and an 8.3% drop in accuracy moving from 32k to 2k context lengths for GPT-4. Finally, we report correlations between clinician rankings and automated natural language generation metrics as a way to rank LLMs without human review. We make MedAlign available under a research data use agreement to enable LLM evaluations on tasks aligned with clinician needs and preferences.
DynImp: Dynamic Imputation for Wearable Sensing Data Through Sensory and Temporal Relatedness
Sep 26, 2022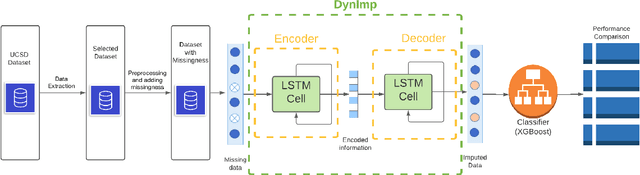
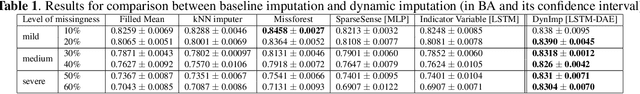
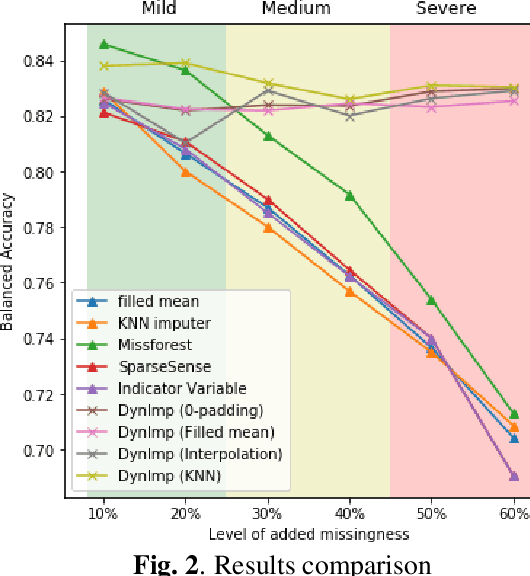
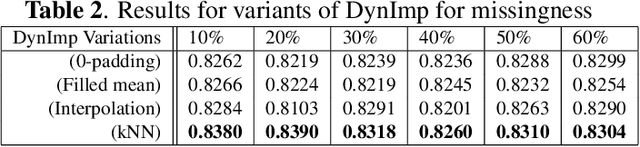
Abstract:In wearable sensing applications, data is inevitable to be irregularly sampled or partially missing, which pose challenges for any downstream application. An unique aspect of wearable data is that it is time-series data and each channel can be correlated to another one, such as x, y, z axis of accelerometer. We argue that traditional methods have rarely made use of both times-series dynamics of the data as well as the relatedness of the features from different sensors. We propose a model, termed as DynImp, to handle different time point's missingness with nearest neighbors along feature axis and then feeding the data into a LSTM-based denoising autoencoder which can reconstruct missingness along the time axis. We experiment the model on the extreme missingness scenario ($>50\%$ missing rate) which has not been widely tested in wearable data. Our experiments on activity recognition show that the method can exploit the multi-modality features from related sensors and also learn from history time-series dynamics to reconstruct the data under extreme missingness.
Density-Aware Personalized Training for Risk Prediction in Imbalanced Medical Data
Jul 29, 2022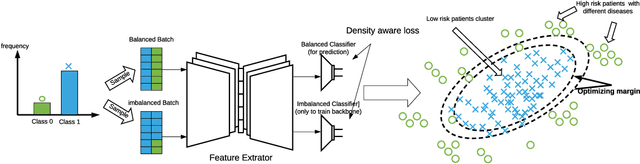

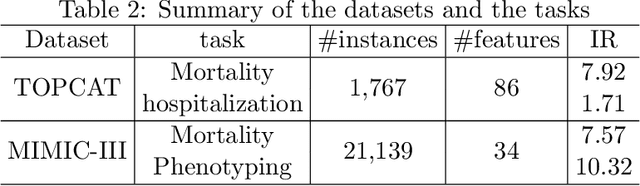
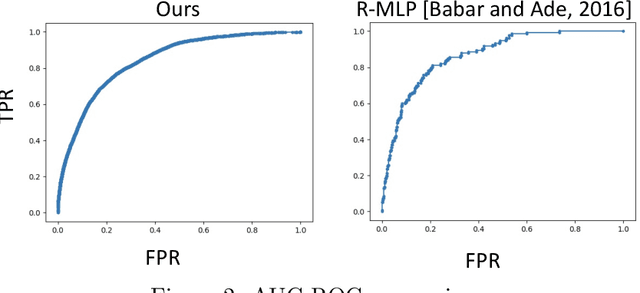
Abstract:Medical events of interest, such as mortality, often happen at a low rate in electronic medical records, as most admitted patients survive. Training models with this imbalance rate (class density discrepancy) may lead to suboptimal prediction. Traditionally this problem is addressed through ad-hoc methods such as resampling or reweighting but performance in many cases is still limited. We propose a framework for training models for this imbalance issue: 1) we first decouple the feature extraction and classification process, adjusting training batches separately for each component to mitigate bias caused by class density discrepancy; 2) we train the network with both a density-aware loss and a learnable cost matrix for misclassifications. We demonstrate our model's improved performance in real-world medical datasets (TOPCAT and MIMIC-III) to show improved AUC-ROC, AUC-PRC, Brier Skill Score compared with the baselines in the domain.
Predicting the meal macronutrient composition from continuous glucose monitors
Jun 23, 2022



Abstract:Sustained high levels of blood glucose in type 2 diabetes (T2DM) can have disastrous long-term health consequences. An essential component of clinical interventions for T2DM is monitoring dietary intake to keep plasma glucose levels within an acceptable range. Yet, current techniques to monitor food intake are time intensive and error prone. To address this issue, we are developing techniques to automatically monitor food intake and the composition of those foods using continuous glucose monitors (CGMs). This article presents the results of a clinical study in which participants consumed nine standardized meals with known macronutrients amounts (carbohydrate, protein, and fat) while wearing a CGM. We built a multitask neural network to estimate the macronutrient composition from the CGM signal, and compared it against a baseline linear regression. The best prediction result comes from our proposed neural network, trained with subject-dependent data, as measured by root mean squared relative error and correlation coefficient. These findings suggest that it is possible to estimate macronutrient composition from CGM signals, opening the possibility to develop automatic techniques to track food intake.
Uncertainty Quantification for Deep Context-Aware Mobile Activity Recognition and Unknown Context Discovery
Mar 03, 2020



Abstract:Activity recognition in wearable computing faces two key challenges: i) activity characteristics may be context-dependent and change under different contexts or situations; ii) unknown contexts and activities may occur from time to time, requiring flexibility and adaptability of the algorithm. We develop a context-aware mixture of deep models termed the {\alpha}-\b{eta} network coupled with uncertainty quantification (UQ) based upon maximum entropy to enhance human activity recognition performance. We improve accuracy and F score by 10% by identifying high-level contexts in a data-driven way to guide model development. In order to ensure training stability, we have used a clustering-based pre-training in both public and in-house datasets, demonstrating improved accuracy through unknown context discovery.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge