Bobak J. Mortazavi
ArterialNet: Reconstructing Arterial Blood Pressure Waveform with Wearable Pulsatile Signals, a Cohort-Aware Approach
Oct 24, 2024



Abstract:Continuous arterial blood pressure (ABP) monitoring is invasive but essential for hemodynamic monitoring. Recent techniques have reconstructed ABP non-invasively using pulsatile signals but produced inaccurate systolic and diastolic blood pressure (SBP and DBP) values and were sensitive to individual variability. ArterialNet integrates generalized pulsatile-to-ABP signal translation and personalized feature extraction using hybrid loss functions and regularization. We validated ArterialNet using the MIMIC-III dataset and achieved a root mean square error (RMSE) of 5.41 mmHg, with at least a 58% lower standard deviation. ArterialNet reconstructed ABP with an RMSE of 7.99 mmHg in remote health scenarios. ArterialNet achieved superior performance in ABP reconstruction and SBP and DBP estimations, with significantly reduced subject variance, demonstrating its potential in remote health settings. We also ablated ArterialNet architecture to investigate the contributions of each component and evaluated its translational impact and robustness by conducting a series of ablations on data quality and availability.
An Efficient Contrastive Unimodal Pretraining Method for EHR Time Series Data
Oct 11, 2024



Abstract:Machine learning has revolutionized the modeling of clinical timeseries data. Using machine learning, a Deep Neural Network (DNN) can be automatically trained to learn a complex mapping of its input features for a desired task. This is particularly valuable in Electronic Health Record (EHR) databases, where patients often spend extended periods in intensive care units (ICUs). Machine learning serves as an efficient method for extract meaningful information. However, many state-of-the-art (SOTA) methods for training DNNs demand substantial volumes of labeled data, posing significant challenges for clinics in terms of cost and time. Self-supervised learning offers an alternative by allowing practitioners to extract valuable insights from data without the need for costly labels. Yet, current SOTA methods often necessitate large data batches to achieve optimal performance, increasing computational demands. This presents a challenge when working with long clinical timeseries data. To address this, we propose an efficient method of contrastive pretraining tailored for long clinical timeseries data. Our approach utilizes an estimator for negative pair comparison, enabling effective feature extraction. We assess the efficacy of our pretraining using standard self-supervised tasks such as linear evaluation and semi-supervised learning. Additionally, our model demonstrates the ability to impute missing measurements, providing clinicians with deeper insights into patient conditions. We demonstrate that our pretraining is capable of achieving better performance as both the size of the model and the size of the measurement vocabulary scale. Finally, we externally validate our model, trained on the MIMIC-III dataset, using the eICU dataset. We demonstrate that our model is capable of learning robust clinical information that is transferable to other clinics.
Density-Aware Personalized Training for Risk Prediction in Imbalanced Medical Data
Jul 29, 2022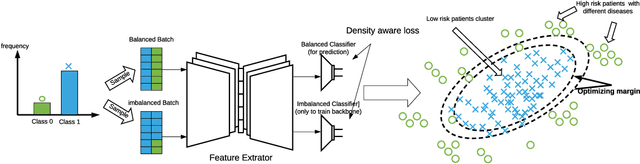

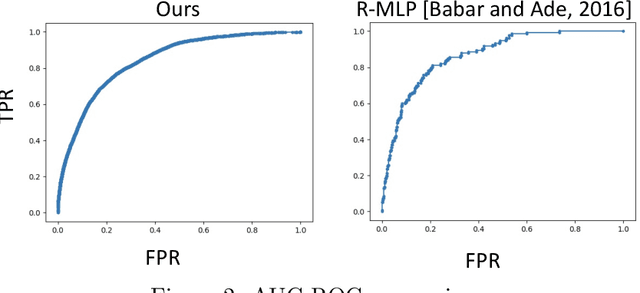
Abstract:Medical events of interest, such as mortality, often happen at a low rate in electronic medical records, as most admitted patients survive. Training models with this imbalance rate (class density discrepancy) may lead to suboptimal prediction. Traditionally this problem is addressed through ad-hoc methods such as resampling or reweighting but performance in many cases is still limited. We propose a framework for training models for this imbalance issue: 1) we first decouple the feature extraction and classification process, adjusting training batches separately for each component to mitigate bias caused by class density discrepancy; 2) we train the network with both a density-aware loss and a learnable cost matrix for misclassifications. We demonstrate our model's improved performance in real-world medical datasets (TOPCAT and MIMIC-III) to show improved AUC-ROC, AUC-PRC, Brier Skill Score compared with the baselines in the domain.
Predicting the meal macronutrient composition from continuous glucose monitors
Jun 23, 2022



Abstract:Sustained high levels of blood glucose in type 2 diabetes (T2DM) can have disastrous long-term health consequences. An essential component of clinical interventions for T2DM is monitoring dietary intake to keep plasma glucose levels within an acceptable range. Yet, current techniques to monitor food intake are time intensive and error prone. To address this issue, we are developing techniques to automatically monitor food intake and the composition of those foods using continuous glucose monitors (CGMs). This article presents the results of a clinical study in which participants consumed nine standardized meals with known macronutrients amounts (carbohydrate, protein, and fat) while wearing a CGM. We built a multitask neural network to estimate the macronutrient composition from the CGM signal, and compared it against a baseline linear regression. The best prediction result comes from our proposed neural network, trained with subject-dependent data, as measured by root mean squared relative error and correlation coefficient. These findings suggest that it is possible to estimate macronutrient composition from CGM signals, opening the possibility to develop automatic techniques to track food intake.
Boosted-SpringDTW for Comprehensive Feature Extraction of Physiological Signals
Jan 11, 2022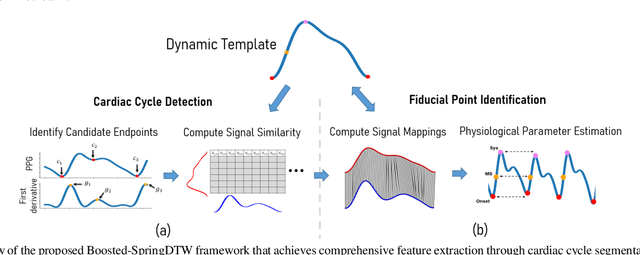



Abstract:Goal: To achieve-high quality comprehensive feature extraction from physiological signals that enables precise physiological parameter estimation despite evolving waveform morphologies. Methods: We propose Boosted-SpringDTW, a probabilistic framework that leverages dynamic time warping (DTW) and minimal domain-specific heuristics to simultaneously segment physiological signals and identify fiducial points that represent cardiac events. An automated dynamic template adapts to evolving waveform morphologies. We validate Boosted-SpringDTW performance with a benchmark PPG dataset whose morphologies include subject- and respiratory-induced variation. Results: Boosted-SpringDTW achieves precision, recall, and F1-scores over 0.96 for identifying fiducial points and mean absolute error values less than 11.41 milliseconds when estimating IBI. Conclusion: Boosted-SpringDTW improves F1-Scores compared to two baseline feature extraction algorithms by 35 percent on average for fiducial point identification and mean percent difference by 16 percent on average for IBI estimation. Significance: Precise hemodynamic parameter estimation with wearable devices enables continuous health monitoring throughout a patients' daily life.
Real-time Mortality Prediction Using MIMIC-IV ICU Data Via Boosted Nonparametric Hazards
Nov 10, 2021
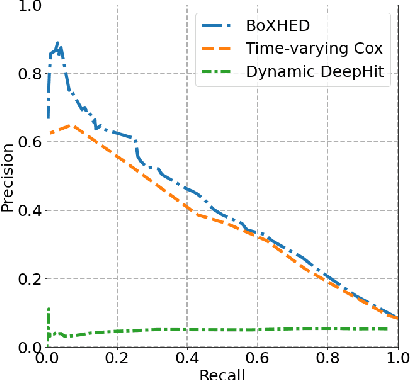
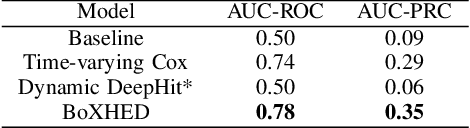
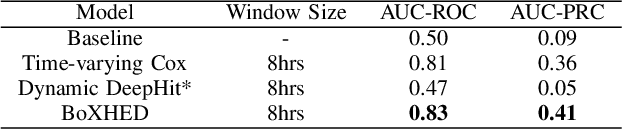
Abstract:Electronic Health Record (EHR) systems provide critical, rich and valuable information at high frequency. One of the most exciting applications of EHR data is in developing a real-time mortality warning system with tools from survival analysis. However, most of the survival analysis methods used recently are based on (semi)parametric models using static covariates. These models do not take advantage of the information conveyed by the time-varying EHR data. In this work, we present an application of a highly scalable survival analysis method, BoXHED 2.0 to develop a real-time in-ICU mortality warning indicator based on the MIMIC IV data set. Importantly, BoXHED can incorporate time-dependent covariates in a fully nonparametric manner and is backed by theory. Our in-ICU mortality model achieves an AUC-PRC of 0.41 and AUC-ROC of 0.83 out of sample, demonstrating the benefit of real-time monitoring.
BoXHED 2.0: Scalable boosting of functional data in survival analysis
Mar 23, 2021



Abstract:Modern applications of survival analysis increasingly involve time-dependent covariates, which constitute a form of functional data. Learning from functional data generally involves repeated evaluations of time integrals which is numerically expensive. In this work we propose a lightweight data preprocessing step that transforms functional data into nonfunctional data. Boosting implementations for nonfunctional data can then be used, whereby the required numerical integration comes for free as part of the training phase. We use this to develop BoXHED 2.0, a quantum leap over the tree-boosted hazard package BoXHED 1.0. BoXHED 2.0 extends BoXHED 1.0 to Aalen's multiplicative intensity model, which covers censoring schemes far beyond right-censoring and also supports recurrent events data. It is also massively scalable because of preprocessing and also because it borrows from the core components of XGBoost. BoXHED 2.0 supports the use of GPUs and multicore CPUs, and is available from GitHub: www.github.com/BoXHED.
Dynamically Extracting Outcome-Specific Problem Lists from Clinical Notes with Guided Multi-Headed Attention
Jul 25, 2020



Abstract:Problem lists are intended to provide clinicians with a relevant summary of patient medical issues and are embedded in many electronic health record systems. Despite their importance, problem lists are often cluttered with resolved or currently irrelevant conditions. In this work, we develop a novel end-to-end framework that first extracts diagnosis and procedure information from clinical notes and subsequently uses the extracted medical problems to predict patient outcomes. This framework is both more performant and more interpretable than existing models used within the domain, achieving an AU-ROC of 0.710 for bounceback readmission and 0.869 for in-hospital mortality occurring after ICU discharge. We identify risk factors for both readmission and mortality outcomes and demonstrate that our framework can be used to develop dynamic problem lists that present clinical problems along with their quantitative importance. We conduct a qualitative user study with medical experts and demonstrate that they view the lists produced by our framework favorably and find them to be a more effective clinical decision support tool than a strong baseline.
Developing Personalized Models of Blood Pressure Estimation from Wearable Sensors Data Using Minimally-trained Domain Adversarial Neural Networks
Jul 24, 2020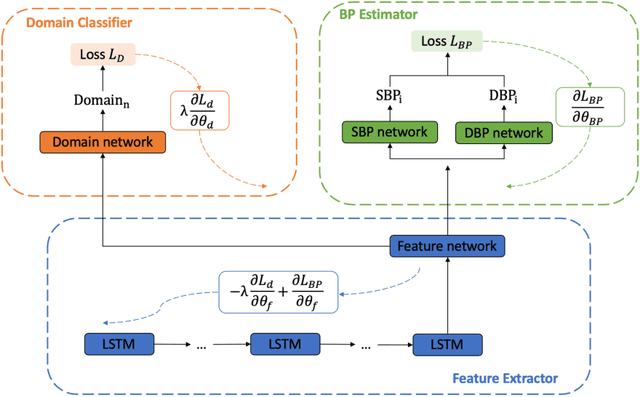
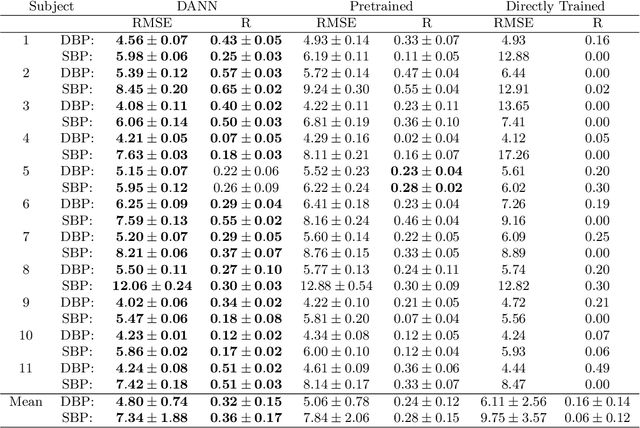
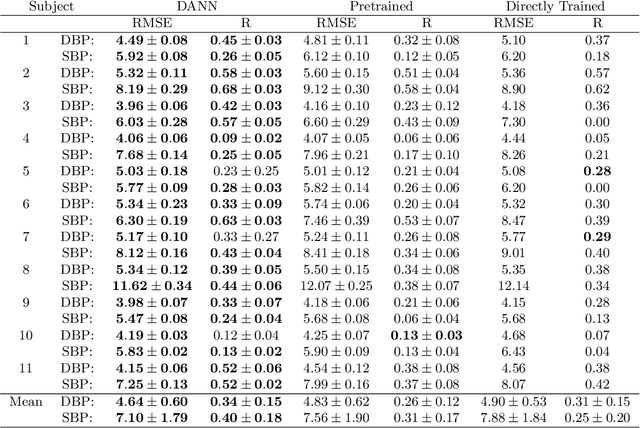
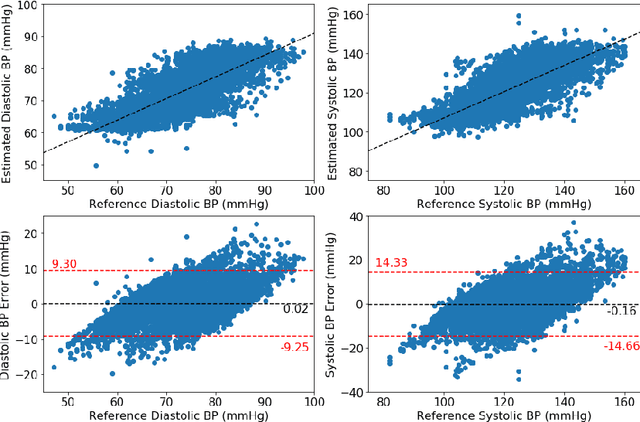
Abstract:Blood pressure monitoring is an essential component of hypertension management and in the prediction of associated comorbidities. Blood pressure is a dynamic vital sign with frequent changes throughout a given day. Capturing blood pressure remotely and frequently (also known as ambulatory blood pressure monitoring) has traditionally been achieved by measuring blood pressure at discrete intervals using an inflatable cuff. However, there is growing interest in developing a cuffless ambulatory blood pressure monitoring system to measure blood pressure continuously. One such approach is by utilizing bioimpedance sensors to build regression models. A practical problem with this approach is that the amount of data required to confidently train such a regression model can be prohibitive. In this paper, we propose the application of the domain-adversarial training neural network (DANN) method on our multitask learning (MTL) blood pressure estimation model, allowing for knowledge transfer between subjects. Our proposed model obtains average root mean square error (RMSE) of $4.80 \pm 0.74$ mmHg for diastolic blood pressure and $7.34 \pm 1.88$ mmHg for systolic blood pressure when using three minutes of training data, $4.64 \pm 0.60$ mmHg and $7.10 \pm 1.79$ respectively when using four minutes of training data, and $4.48 \pm 0.57$ mmHg and $6.79 \pm 1.70$ respectively when using five minutes of training data. DANN improves training with minimal data in comparison to both directly training and to training with a pretrained model from another subject, decreasing RMSE by $0.19$ to $0.26$ mmHg (diastolic) and by $0.46$ to $0.67$ mmHg (systolic) in comparison to the best baseline models. We observe that four minutes of training data is the minimum requirement for our framework to exceed ISO standards within this cohort of patients.
BoXHED: Boosted eXact Hazard Estimator with Dynamic covariates
Jun 26, 2020

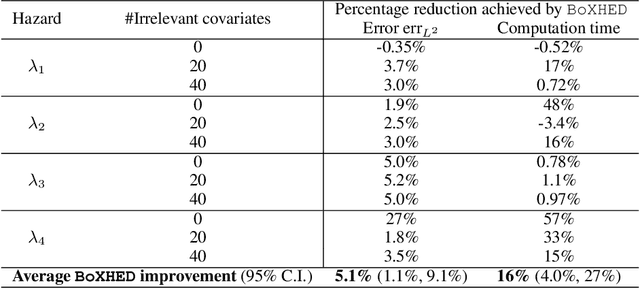
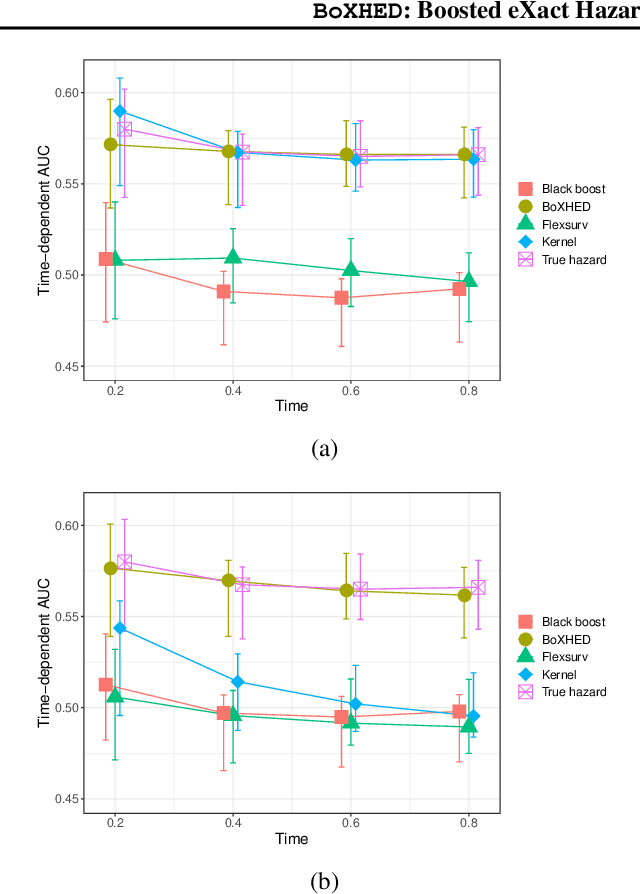
Abstract:The proliferation of medical monitoring devices makes it possible to track health vitals at high frequency, enabling the development of dynamic health risk scores that change with the underlying readings. Survival analysis, in particular hazard estimation, is well-suited to analyzing this stream of data to predict disease onset as a function of the time-varying vitals. This paper introduces the software package BoXHED (pronounced 'box-head') for nonparametrically estimating hazard functions via gradient boosting. BoXHED 1.0 is a novel tree-based implementation of the generic estimator proposed in Lee, Chen, Ishwaran (2017), which was designed for handling time-dependent covariates in a fully nonparametric manner. BoXHED is also the first publicly available software implementation for Lee, Chen, Ishwaran (2017). Applying BoXHED to cardiovascular disease onset data from the Framingham Heart Study reveals novel interaction effects among known risk factors, potentially resolving an open question in clinical literature.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge