Ryan King
NREL
An Efficient Contrastive Unimodal Pretraining Method for EHR Time Series Data
Oct 11, 2024



Abstract:Machine learning has revolutionized the modeling of clinical timeseries data. Using machine learning, a Deep Neural Network (DNN) can be automatically trained to learn a complex mapping of its input features for a desired task. This is particularly valuable in Electronic Health Record (EHR) databases, where patients often spend extended periods in intensive care units (ICUs). Machine learning serves as an efficient method for extract meaningful information. However, many state-of-the-art (SOTA) methods for training DNNs demand substantial volumes of labeled data, posing significant challenges for clinics in terms of cost and time. Self-supervised learning offers an alternative by allowing practitioners to extract valuable insights from data without the need for costly labels. Yet, current SOTA methods often necessitate large data batches to achieve optimal performance, increasing computational demands. This presents a challenge when working with long clinical timeseries data. To address this, we propose an efficient method of contrastive pretraining tailored for long clinical timeseries data. Our approach utilizes an estimator for negative pair comparison, enabling effective feature extraction. We assess the efficacy of our pretraining using standard self-supervised tasks such as linear evaluation and semi-supervised learning. Additionally, our model demonstrates the ability to impute missing measurements, providing clinicians with deeper insights into patient conditions. We demonstrate that our pretraining is capable of achieving better performance as both the size of the model and the size of the measurement vocabulary scale. Finally, we externally validate our model, trained on the MIMIC-III dataset, using the eICU dataset. We demonstrate that our model is capable of learning robust clinical information that is transferable to other clinics.
Multimodal Pretraining of Medical Time Series and Notes
Dec 11, 2023Abstract:Within the intensive care unit (ICU), a wealth of patient data, including clinical measurements and clinical notes, is readily available. This data is a valuable resource for comprehending patient health and informing medical decisions, but it also contains many challenges in analysis. Deep learning models show promise in extracting meaningful patterns, but they require extensive labeled data, a challenge in critical care. To address this, we propose a novel approach employing self-supervised pretraining, focusing on the alignment of clinical measurements and notes. Our approach combines contrastive and masked token prediction tasks during pretraining. Semi-supervised experiments on the MIMIC-III dataset demonstrate the effectiveness of our self-supervised pretraining. In downstream tasks, including in-hospital mortality prediction and phenotyping, our pretrained model outperforms baselines in settings where only a fraction of the data is labeled, emphasizing its ability to enhance ICU data analysis. Notably, our method excels in situations where very few labels are available, as evidenced by an increase in the AUC-ROC for in-hospital mortality by 0.17 and in AUC-PR for phenotyping by 0.1 when only 1% of labels are accessible. This work advances self-supervised learning in the healthcare domain, optimizing clinical insights from abundant yet challenging ICU data.
Growing Representation Learning
Oct 17, 2021



Abstract:Machine learning continues to grow in popularity due to its ability to learn increasingly complex tasks. However, for many supervised models, the shift in a data distribution or the appearance of a new event can result in a severe decrease in model performance. Retraining a model from scratch with updated data can be resource intensive or impossible depending on the constraints placed on an organization or system. Continual learning methods attempt to adapt models to new classes instead of retraining. However, many of these methods do not have a detection method for new classes or make assumptions about the distribution of classes. In this paper, we develop an attention based Gaussian Mixture, called GMAT, that learns interpretable representations of data with or without labels. We incorporate this method with existing Neural Architecture Search techniques to develop an algorithm for detection new events for an optimal number of representations through an iterative process of training a growing. We show that our method is capable learning new representations of data without labels or assumptions about the distributions of labels. We additionally develop a method that allows our model to utilize labels to more accurately develop representations. Lastly, we show that our method can avoid catastrophic forgetting by replaying samples from learned representations.
A block-random algorithm for learning on distributed, heterogeneous data
Feb 28, 2019



Abstract:Most deep learning models are based on deep neural networks with multiple layers between input and output. The parameters defining these layers are initialized using random values and are "learned" from data, typically using stochastic gradient descent based algorithms. These algorithms rely on data being randomly shuffled before optimization. The randomization of the data prior to processing in batches that is formally required for stochastic gradient descent algorithm to effectively derive a useful deep learning model is expected to be prohibitively expensive for in situ model training because of the resulting data communications across the processor nodes. We show that the stochastic gradient descent (SGD) algorithm can still make useful progress if the batches are defined on a per-processor basis and processed in random order even though (i) the batches are constructed from data samples from a single class or specific flow region, and (ii) the overall data samples are heterogeneous. We present block-random gradient descent, a new algorithm that works on distributed, heterogeneous data without having to pre-shuffle. This algorithm enables in situ learning for exascale simulations. The performance of this algorithm is demonstrated on a set of benchmark classification models and the construction of a subgrid scale large eddy simulations (LES) model for turbulent channel flow using a data model similar to that which will be encountered in exascale simulation.
From Deep to Physics-Informed Learning of Turbulence: Diagnostics
Oct 16, 2018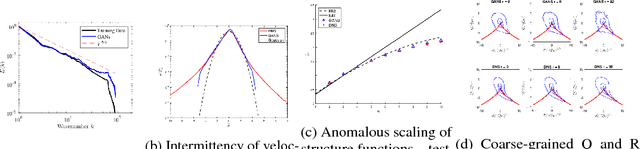
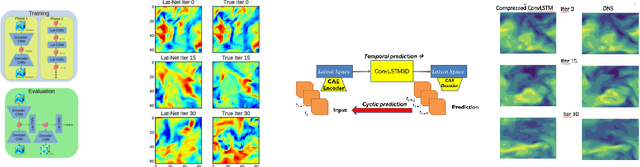
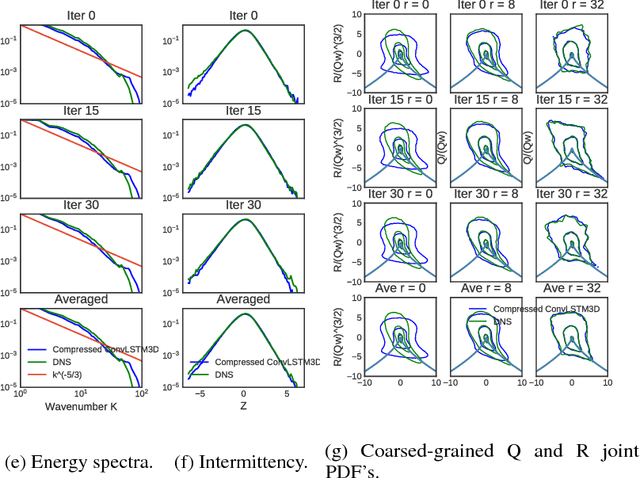
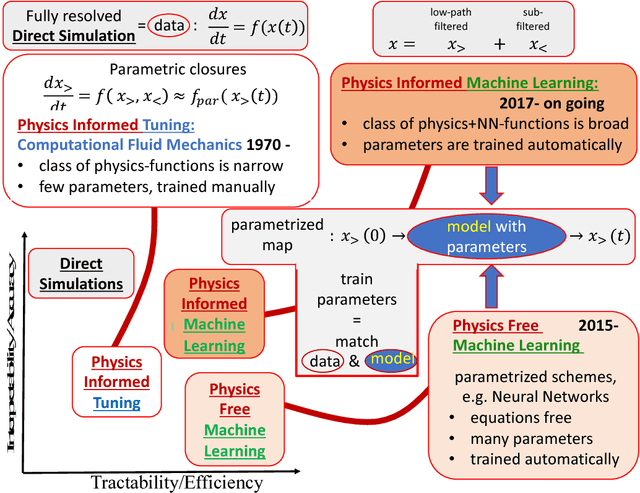
Abstract:We describe physical tests validating progress made toward acceleration and automation of hydrodynamic codes in the regime of developed turbulence by two {\bf Deep Learning} (DL) Neural Network (NN) schemes trained on {\bf Direct Numerical Simulations} of turbulence. Even the bare DL solutions, which do not take into account any physics of turbulence explicitly, are impressively good overall when it comes to qualitative description of important features of turbulence. However, the early tests have also uncovered some caveats of the DL approaches. We observe that the static DL scheme, implementing Convolutional GAN and trained on spatial snapshots of turbulence, fails to reproduce intermittency of turbulent fluctuations at small scales and details of the turbulence geometry at large scales. We show that the dynamic NN scheme, LAT-NET, trained on a temporal sequence of turbulence snapshots is capable to correct for the small-scale caveat of the static NN. We suggest a path forward towards improving reproducibility of the large-scale geometry of turbulence with NN.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge