Syed Jamal Safdar Gardezi
Department of Radiology, University of Wisconsin School of Medicine & Public Health, Madison, WI, USA
3D Nephrographic Image Synthesis in CT Urography with the Diffusion Model and Swin Transformer
Feb 26, 2025Abstract:Purpose: This study aims to develop and validate a method for synthesizing 3D nephrographic phase images in CT urography (CTU) examinations using a diffusion model integrated with a Swin Transformer-based deep learning approach. Materials and Methods: This retrospective study was approved by the local Institutional Review Board. A dataset comprising 327 patients who underwent three-phase CTU (mean $\pm$ SD age, 63 $\pm$ 15 years; 174 males, 153 females) was curated for deep learning model development. The three phases for each patient were aligned with an affine registration algorithm. A custom deep learning model coined dsSNICT (diffusion model with a Swin transformer for synthetic nephrographic phase images in CT) was developed and implemented to synthesize the nephrographic images. Performance was assessed using Peak Signal-to-Noise Ratio (PSNR), Structural Similarity Index (SSIM), Mean Absolute Error (MAE), and Fr\'{e}chet Video Distance (FVD). Qualitative evaluation by two fellowship-trained abdominal radiologists was performed. Results: The synthetic nephrographic images generated by our proposed approach achieved high PSNR (26.3 $\pm$ 4.4 dB), SSIM (0.84 $\pm$ 0.069), MAE (12.74 $\pm$ 5.22 HU), and FVD (1323). Two radiologists provided average scores of 3.5 for real images and 3.4 for synthetic images (P-value = 0.5) on a Likert scale of 1-5, indicating that our synthetic images closely resemble real images. Conclusion: The proposed approach effectively synthesizes high-quality 3D nephrographic phase images. This model can be used to reduce radiation dose in CTU by 33.3\% without compromising image quality, which thereby enhances the safety and diagnostic utility of CT urography.
Merlin: A Vision Language Foundation Model for 3D Computed Tomography
Jun 10, 2024



Abstract:Over 85 million computed tomography (CT) scans are performed annually in the US, of which approximately one quarter focus on the abdomen. Given the current radiologist shortage, there is a large impetus to use artificial intelligence to alleviate the burden of interpreting these complex imaging studies. Prior state-of-the-art approaches for automated medical image interpretation leverage vision language models (VLMs). However, current medical VLMs are generally limited to 2D images and short reports, and do not leverage electronic health record (EHR) data for supervision. We introduce Merlin - a 3D VLM that we train using paired CT scans (6+ million images from 15,331 CTs), EHR diagnosis codes (1.8+ million codes), and radiology reports (6+ million tokens). We evaluate Merlin on 6 task types and 752 individual tasks. The non-adapted (off-the-shelf) tasks include zero-shot findings classification (31 findings), phenotype classification (692 phenotypes), and zero-shot cross-modal retrieval (image to findings and image to impressions), while model adapted tasks include 5-year disease prediction (6 diseases), radiology report generation, and 3D semantic segmentation (20 organs). We perform internal validation on a test set of 5,137 CTs, and external validation on 7,000 clinical CTs and on two public CT datasets (VerSe, TotalSegmentator). Beyond these clinically-relevant evaluations, we assess the efficacy of various network architectures and training strategies to depict that Merlin has favorable performance to existing task-specific baselines. We derive data scaling laws to empirically assess training data needs for requisite downstream task performance. Furthermore, unlike conventional VLMs that require hundreds of GPUs for training, we perform all training on a single GPU.
Automatic Segmentation of the Kidneys and Cystic Renal Lesions on Non-Contrast CT Using a Convolutional Neural Network
May 14, 2024



Abstract:Objective: Automated segmentation tools are useful for calculating kidney volumes rapidly and accurately. Furthermore, these tools have the power to facilitate large-scale image-based artificial intelligence projects by generating input labels, such as for image registration algorithms. Prior automated segmentation models have largely ignored non-contrast computed tomography (CT) imaging. This work aims to implement and train a deep learning (DL) model to segment the kidneys and cystic renal lesions (CRLs) from non-contrast CT scans. Methods: Manual segmentation of the kidneys and CRLs was performed on 150 non-contrast abdominal CT scans. The data were divided into an 80/20 train/test split and a deep learning (DL) model was trained to segment the kidneys and CRLs. Various scoring metrics were used to assess model performance, including the Dice Similarity Coefficient (DSC), Jaccard Index (JI), and absolute and percent error kidney volume and lesion volume. Bland-Altman (B-A) analysis was performed to compare manual versus DL-based kidney volumes. Results: The DL model achieved a median kidney DSC of 0.934, median CRL DSC of 0.711, and total median study DSC of 0.823. Average volume errors were 0.9% for renal parenchyma, 37.0% for CRLs, and 2.2% overall. B-A analysis demonstrated that DL-based volumes tended to be greater than manual volumes, with a mean bias of +3.0 ml (+/- 2 SD of +/- 50.2 ml). Conclusion: A deep learning model trained to segment kidneys and cystic renal lesions on non-contrast CT examinations was able to provide highly accurate segmentations, with a median kidney Dice Similarity Coefficient of 0.934. Keywords: deep learning; kidney segmentation; artificial intelligence; convolutional neural networks.
ResNCT: A Deep Learning Model for the Synthesis of Nephrographic Phase Images in CT Urography
May 07, 2024


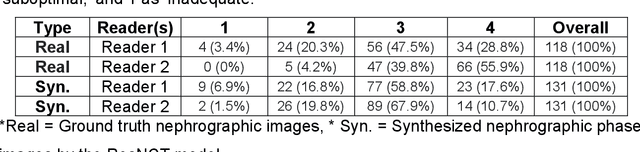
Abstract:Purpose: To develop and evaluate a transformer-based deep learning model for the synthesis of nephrographic phase images in CT urography (CTU) examinations from the unenhanced and urographic phases. Materials and Methods: This retrospective study was approved by the local Institutional Review Board. A dataset of 119 patients (mean $\pm$ SD age, 65 $\pm$ 12 years; 75/44 males/females) with three-phase CT urography studies was curated for deep learning model development. The three phases for each patient were aligned with an affine registration algorithm. A custom model, coined Residual transformer model for Nephrographic phase CT image synthesis (ResNCT), was developed and implemented with paired inputs of non-contrast and urographic sets of images trained to produce the nephrographic phase images, that were compared with the corresponding ground truth nephrographic phase images. The synthesized images were evaluated with multiple performance metrics, including peak signal to noise ratio (PSNR), structural similarity index (SSIM), normalized cross correlation coefficient (NCC), mean absolute error (MAE), and root mean squared error (RMSE). Results: The ResNCT model successfully generated synthetic nephrographic images from non-contrast and urographic image inputs. With respect to ground truth nephrographic phase images, the images synthesized by the model achieved high PSNR (27.8 $\pm$ 2.7 dB), SSIM (0.88 $\pm$ 0.05), and NCC (0.98 $\pm$ 0.02), and low MAE (0.02 $\pm$ 0.005) and RMSE (0.042 $\pm$ 0.016). Conclusion: The ResNCT model synthesized nephrographic phase CT images with high similarity to ground truth images. The ResNCT model provides a means of eliminating the acquisition of the nephrographic phase with a resultant 33% reduction in radiation dose for CTU examinations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge